A universal SNP and small-indel variant caller using deep neural networks
Background
Genes determine Biological functions, and mutants or alleles(one of two or more alternative forms of a gene that arise by mutation and are found at the same place on a chromosome) of those genes determine differences within a function. Determining novel alleles is very important in understanding the genetic variation within a species. For example, different alleles of the gene OCA2 determine the colour of pupils. All animals receive one copy of each gene from each of their parents. Mutations of a gene are classified as either homozygous (both copies are the same) or heterozygous (the two copies are different).
Next-generation sequencing is a prevalent technique for sequencing or reading DNA. Since all genes are encoded as DNA, sequencing is an essential tool for understanding genes. Next-generation sequencing works by reading short sections of DNA of length k, called k-means, and then piecing them together or aligning them to a reference genome. Next-generation sequencing is relatively fast and inexpensive, although it can randomly misidentify some nucleotides, introducing errors. However, NGS reading is errorful and arises from a complex error process depending on various factors.
The process of variant calling is determining novel alleles from sequencing data (typically next-generation sequencing data). Some significant alleles only differ from the "standard" version of a gene by only a single base pair, such as the mutation which causes multiple sclerosis. Therefore it is crucial to accurately call single nucleotide swaps/polymorphisms (SNPs), insertions, and deletions (indels). Calling SNPs and small indels are technically challenging since it requires a program to distinguish between genuinely novel mutations and errors in the sequencing data.
Previous approaches usually involved using various statistical techniques. A widely used one is GATK. GATK uses a combination of logistic regression, hidden Markov models, naive Bayes classification, and Gaussian mixture models to perform the process [2]. However, these methods have their weaknesses as some assumptions do not hold (i.e., independence assumptions). In addition, given that GATK testing has focused primarily on human whole-genome data sequenced using Illumina technology, it is not easily generalizable to different types of data, organisms, and experimental designs/sequencing technologies [3].
This paper aims to solve the problem of calling SNPs and small indels using a convolutional neural net by casting the reads as images and classifying whether they contain a mutation. It introduces a variant caller called "DeepVariant", which requires no specialized knowledge, but performs better than previous state-of-art methods.
Overview
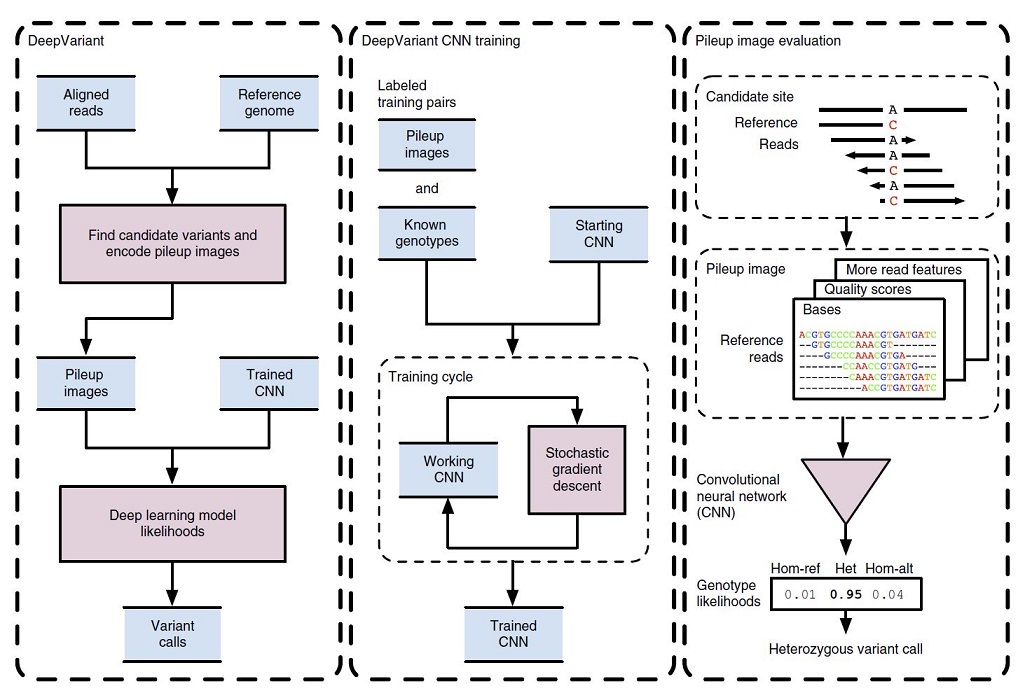
In Figure 1, the DeepVariant workflow overview is illustrated.
Initially, the NGS reads aligned to a reference genome which are then scanned for candidate variants which are different sites from the reference genome. The read and reference data are encoded as an image for each candidate variant site. Then, the trained CNN can compute the genotype likelihoods, (heterozygous or homozygous) for each of the candidate variants (figure1, left box).
To train the CNN for image classification purposes, the DeepVariant machinery makes pileup images for a labeled sample with known genotypes. These labeled images and known genotypes are provided to CNN for training, and a stochastic gradient descent algorithm is used to optimize the CNN parameters to maximize genotype prediction accuracy. After the convergence of the model, the final model is frozen to use for calling mutations for other image classification tests (figure1, middle box).
For example, in figure 1 (right box), the reference and read bases are encoded into a pileup image at a candidate variant site. The CNN using this encoded image computes the genotype likelihoods for the three diploid genotype states of homozygous reference (hom-ref), heterozygous (het) or homozygous alternate (hom-alt). In this example, a heterozygous variant call is emitted, as the most probable genotype here is “het”.
Preprocessing
Before the sequencing reads can be fed into the classifier, they must be pre-processed. There are many pre-processing steps that are necessary for this algorithm. These steps represent the real novelty in this technique by transforming the data to allow us to use more common neural network architectures for classification. The pre-processing of the data can be broken into three phases: the realignment of reads, finding candidate variants and creating the candidate variants' images.
The realignment of the pre-processing reads phase is essential to ensure the sequences can be adequately compared to the reference sequences. First, the sequences are aligned to a reference sequence. Reads that align poorly are grouped with other reads around them to build that section, or haplotype, from scratch. If there is strong evidence that the new version of the haplotype fits the reads well, the reads are re-aligned. This process updates the CIGAR (Compact Idiosyncratic Gapped Alignment Report) string to represent a sequence's alignment to a reference for each read.
Once the reads are correctly aligned, the algorithm then proceeds to find candidate variants, regions in the DNA sequence containing variants. It is these candidate variants that will eventually be passed as input to the neural network. To find these, we need to consider each position in the reference sequence independently. Any unusable reads are filtered at this point. This includes reads that are not appropriately aligned, marked as duplicates, those that fail vendor quality checks, or whose mapping quality is less than ten. For each site in the genome, we collect all the remaining reads that overlap that site. The corresponding allele aligned to that site is then determined by decoding the CIGAR string, which was updated in each read's realignment phase. The alleles are then classified into one of four categories: reference-matching base, reference-mismatching base, insertion with a specific sequence, or deletion with a specific length, and the number of occurrences of each distinct allele across all reads is counted. Read bases are only included as potential alleles if each base in the allele has a quality score of at least 10.
The last phase of pre-processing is to convert these candidate variants into images representing the data with candidate variants identified. This allows for the use of well established convolutional neural networks for image classification for this technical problem. Each color channel is used to store a different piece of information about a candidate variant. The red channel encodes which base we have (A, G, C, or T) by mapping each base to a particular value. The quality of the read is mapped to the green color channel.
Moreover, the blue channel encodes whether or not the reference is on the positive strand of the DNA. Each row of the image represents a read, and each column represents a particular base in that read. The reference strand is repeated for the first five rows of the encoded image to maintain its information after a 5x5 convolution is applied. With the data pre-processing complete, the images can then be passed into the neural network for classification.
Neural Network
The neural network used is a convolutional neural network. Although the full network architecture is not revealed in the paper, there are several details which we can discuss. The architecture of the network is an input layer attached to an adapted Inception v2 ImageNet model with nine partitions. The inception v2 model in particular uses a series of CNNs. One interesting aspect about the Inception model is that rather than optimizing a series of hyperparameters in order to determine the most optimal parameter configuration, Inception instead concatenates a series of different sizes of filters on the same layer, which acts to learn the best architecture out of these concatenated filters. The input layer takes as input the images representing the candidate variants and rescales them to 299x299 pixels. The output layer is a three-class Softmax layer initialized with Gaussian random weights with a standard deviation of 0.001. This final layer is fully connected to the previous layer. The three classes are the homozygous reference (meaning it is not a variant), heterozygous variant, and homozygous variant. The candidate variant is classified into the class with the highest probability. The model is trained using stochastic gradient descent with a weight decay of 0.00004. The training was done in mini-batches, each with 32 images, using a root mean squared (RMS) decay of 0.9. For the multiple sequencing technologies experiments, a single model was trained with a learning rate of 0.0015 and momentum 0.8 for 250,000 update steps. For all other experiments, multiple models were trained, and the one with the highest accuracy on the training set was chosen as the final model. The multiple models stem from using each combination of the possible parameter values for the learning rate (0.00095, 0.001, 0.0015) and momentum (0.8, 0.85, 0.9). These models were trained for 80 hours, or until the training accuracy converged.
Results
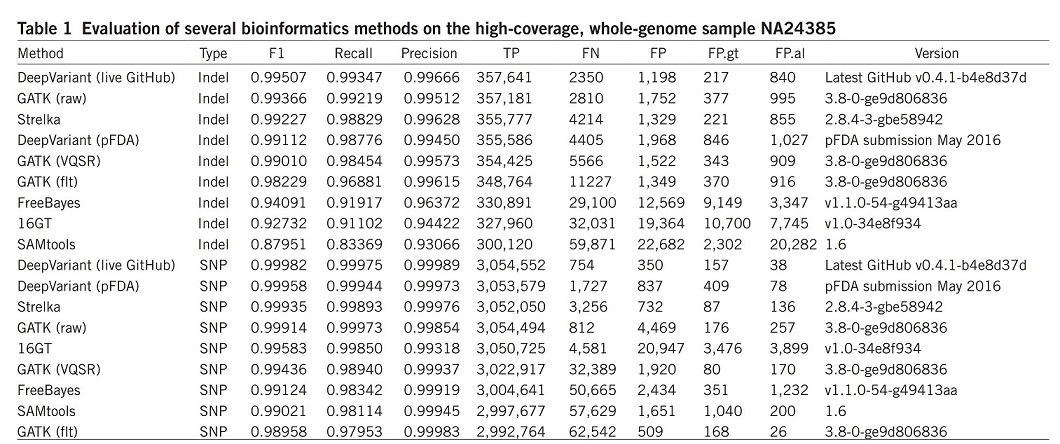
DeepVariant was trained using data available from the CEPH (Centre d’Etude du Polymorphism Humain) female sample NA12878 and was evaluated on the unseen Ashkenazi male sample NA24385. The results were compared with other most commonly used bioinformatics methods, such as the GATK, FreeBayes22, SAMtools23, 16GT24 and Strelka25 (Table 1). For better comparison, the overall accuracy (F1), recall, precision, and numbers of true positives (TP), false negatives (FN) and false positives (FP) are illustrated over the whole genome.
DeepVariant showed the highest accuracy and more than 50% fewer errors per genome compared to the next best algorithm.
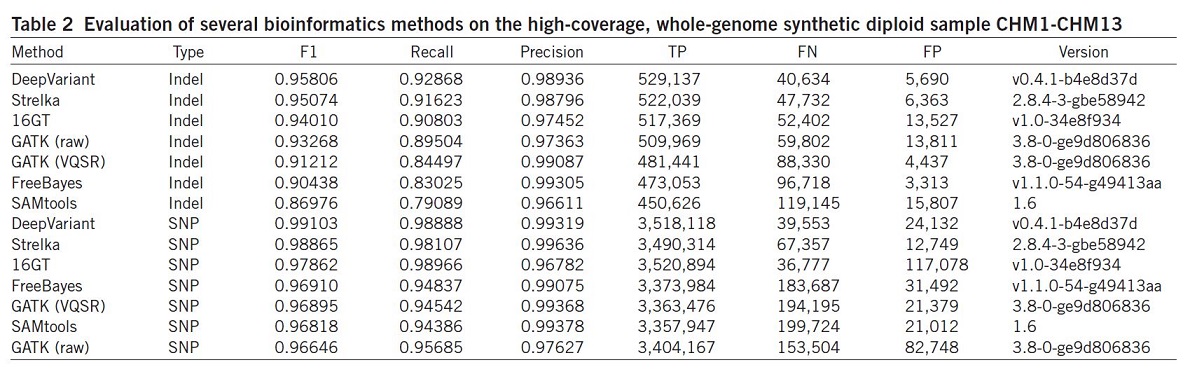
They also evaluated the same set of algorithms using the synthetic diploid sample CHM1-CHM1326 (Table 2).
Results illustrated that the DeepVariant method outperformed all other algorithms for variant calling (SNP and indel) and showed the highest accuracy in terms of F1, Recall, precision and TP.
Conclusion
This endeavor to further advance a data-centric approach to understanding the gene sequence illustrates the advantages of deep learning over humans. With billions of DNA base pairs, no humans can digest that amount of gene expressions. In the past, computational techniques are unfeasible due to the lack of computing power, but in the 21st century, it seems that machine learning is the way to go for molecular biology.
DeepVariant’s strong performance on human data proves that deep learning is a promising technique for variant calling. Perhaps the most exciting feature of DeepVariant is its simplicity. Unlike other states of the art variant callers, DeepVariant does not know the sequencing technologies that create the reads or even the biological processes that introduce mutations. It simplifies the problem of variant calling to preprocessing the reads and training a generic deep learning model. It also suggests that DeepVariant could be significantly improved by tailoring the preprocessing to specific sequencing technologies and developing a dedicated CNN architecture for the reads, rather than casting them as images.
Online Methods
Besides the main topic, the paper also introduced some valuable online methods. These methods have cross intersection with the main topic, although not implicitly, they really showed how some of the other tasks can be tackled using efficient methods.
Haplotype-aware realignment of reads
Mapped reads are preprocessed using an error-tolerant, local De-Bruijn-graph-based read assembly procedure. Candidate windows across the genome are selected for reassembly by looking for any evidence of possible genetic variation. The selection criteria for a candidate window are very permissive so that true variation is unlikely to be missed.
Finding candidate variants
Creating images around candidate variants
Deep learning
Deep Variant inference client and allele merging
Critique and Discussion
The paper presents an attractive method for solving a significant problem. Building "images" of reads and running them through a generic image classification CNN seems like a strange approach, and, interestingly, it works well. The most significant issues with the paper are the lack of specific information about how the methods. Some extra information is included in the supplementary material, but there are still some significant gaps. In particular:
1. What is the structure of the neural net? How many layers, and what sizes? The paper for ConvNet does not have this information. We suspect that this might be a trade secret that Google is protecting.
2. How is the realignment step implemented? The paper mentions that it uses a "De-Bruijn-graph-based read assembly procedure" to realign reads to a new haplotype. It is a non-standard step in most genomics workflows, yet the paper does not describe how they do the realignment or build the haplotypes.
3. How did they settle on the image construction algorithm? The authors provide pseudocode for the construction of pileup images, but they do not describe how to make decisions. For instance, the color values for different base pairs are not evenly spaced. Also, the image begins with five rows of the reference genome.
One thing we appreciated about the paper was their commentary on future developments. The authors clarify that this approach can be improved on and provide specific ideas for the next steps.
Overall, the paper presents an interesting idea with strong results but lacks detail in some vital implementation pieces.
4. The topic of this project is good, but we need more details on the algorithm. In the neural network part, the details are not enough; authors should provide a figure to explain better how the model works and the model's structure. Otherwise, we cannot understand how the model works. When we preprocess the data if different data have different lengths, shall we add more information or drop some information to match?
5. Particularly, which package did the researchers use to perform this analysis? Different packages of deep learning can have different accuracy and efficiency while making predictions on this data set.
Further studies on DeepVariant have shown that it is a framework with great potential and sets the medical standard genetics field.
Another good follow up works can be seen here [1]
6. It was mentioned that part of the network used is an "adapted Inception v2 ImageNet model" - does this mean that it used an Inception v2 model that was trained on ImageNet? This is not clear, but if this is the case, then why is this useful? Why would the features that are extracted for an image be useful for genomics? Did they try using a model that was not trained? Also, they describe the preprocessing method used but were there any alternatives that they considered?
7. A more extensive discussion on the "Neural Network" section can be given. For example, the paragraph's last sentence says that "Models are trained for 80 hours, or until the training accuracy converged." This sentence implies that if the training accuracy does converge, it usually takes less than 80 hours. More exciting data can thus be presented about the training accuracy converging time. How often do the models converge? How long does it take for the models to converge on average? Moreover, why is the number 80 chosen here? Is it a random upper bound set by people to bound the runtime of the model training, or is the number 80 carefully chosen so that it is twice/three times/ten times the average training accuracy converging time?
8. It would be more convincing if the author could provide more detail on the structure of the neural network.
9. It is clear that this is a very thoroughly written paper with substantial comparison results and computation numbers to back up the testing. However, simply because the implementation link is given in the paper, there still lacks information regarding the structure of the model. It is also structurally missing a conclusion section to complete a summary on the overall conclusion and results comparison to give a final conclusion to the efficacy of the proposed model.
10. It would be interesting to see how the model would behave if we incorporate transformers to the model, this has been on reason Alpha Fold 2 was so successful.
11. The result illustration is hard to interpret, it would be nicer if explanations can be added. Lacking details on neural networks used. How was mathematical calculations done on prediction?
12. It would be better if the author could provide a list of other potential networks that could be used to address the problem and a comparison between them.
13. It is very interesting to see that a part of the data preprocessing is to create images from the data as one would normally expect you to simply feed the data itself into the network. By converting to image data does this help the network classify it better or would simply feeding forward the unconverted data be better? This is interesting as at the end of the day an image is still just numerical data so would one representation hold more value over the other?
14. The author opened the door to machine learning solutions in molecular biology by solving genetic problems through data methods. But the details of the neural network training parameters still needs to be clarified. In addition, it is worthy of attention and further explanation about the results of prediction accuracy, under what environment, and how to compare with human predictions.
References
[1] Hartwell, L.H. et. al. Genetics: From Genes to Genomes. (McGraw-Hill Ryerson, 2014).
[2] Poplin, R. et. al. A universal SNP and small-indel variant caller using deep neural networks. Nature Biotechnology 36, 983-987 (2018).