stat841f14: Difference between revisions
| Line 524: | Line 524: | ||
:<math>D^X_{n*n} = \begin{bmatrix} d_11\\d_12\\w_3\\...\\w_d \end{bmatrix}</math> | :<math>D^X_{n*n} = \begin{bmatrix} d_11\\d_12\\w_3\\...\\w_d \end{bmatrix}</math> | ||
=====Theorem===== | =====Theorem===== | ||
If <math>D^(X)</math> is a distance matrix and if <math>K=-\frac{1}{2}HD^(X)H</math><br /> | If <math>D^{(X)}</math> is a distance matrix and if <math>K=-\frac{1}{2}HD^{(X)}H</math><br /> | ||
Then <math>D^(X)</math> is Euclidean iff <math>K</math> is Positive Semi-Definite | Then <math>D^{(X)}</math> is Euclidean iff <math>K</math> is Positive Semi-Definite<br /> | ||
Where <math>H=I-\frac{1}{n}\bold{1}^T\bold{1} </math> and <math>\bold{1}=([1,1,...,1]_{1\times n})^T</math> | |||
=====Equivalence of MDS and PCA===== | =====Equivalence of MDS and PCA===== | ||
Revision as of 21:15, 23 September 2014
Editor Sign Up
Data Visualization (Fall 2014)
Principal Components Analysis (PCA) (Lecture: Sep. 10, 2014)
Introduction
Principal Component Analysis (PCA), first invented by Karl Pearson in 1901, is a statistical technique for data analysis. Its main purpose is to reduce the dimensionality of the data.
Suppose there is a set of data points in a d-dimensional space. PCA’s goal is to find a linear subspace with lower dimensionality p (p [math]\displaystyle{ \leq }[/math] d), such that maximum variance is retained in this lower-dimensional space. The linear subspace can be specified by d orthogonal vectors, such as u1, u2 ... ud which form a new coordinate system, called 'principle components'.In other words, PCA aims to reduce the dimensionality of the data, while preserving its information (or minimizing the loss of information). Information comes from variation. For example, if all data points have the same value along one dimension, that dimension does not carry any information. So, to preserve information, the subspace need to contain components (or dimensions) along which, data has its most variability. However, finding a linear subspace with lower dimensionality which include all data points is not possible in practical problems and loss of information is inevitable. But, we try to reduce this loss and capture most of the features of data.
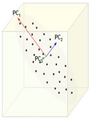
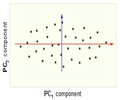
Figure below, demonstrates an example of PCA. Data is transformed from original 3D space to 2D coordinate system where each coordiante is a principal component.
-
Data in original Space
-
Extraction of the principal components
-
Reduction of dimensionality by keeping the first two principal components and removing the third one
PCA Plot example
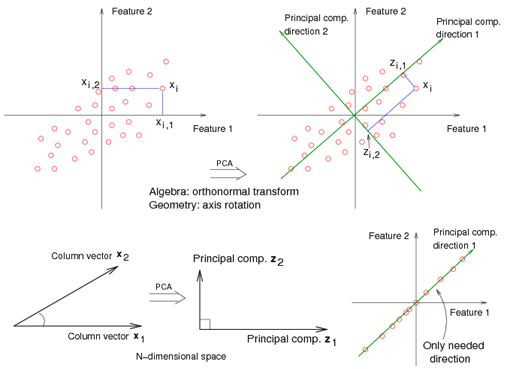
For example<ref> https://onlinecourses.science.psu.edu/stat857/sites/onlinecourses.science.psu.edu.stat857/files/lesson05/PCA_plot.gif </ref>, in the top left corner of the image below, the point [math]\displaystyle{ x_1 }[/math] showed in two-dimensional and it's coordinates are [math]\displaystyle{ (x_1, 1) }[/math] and [math]\displaystyle{ (x_1, 2) }[/math]
All the red points in the plot represented by their projected values on the two original coordinators (Feature 1, Feature 2).
In PCA technique, it uses new coordinators (showed in green). As coordinate system changed, the points also are shown by their new pair of coordinate values for example [math]\displaystyle{ (z_1, 1) }[/math] and [math]\displaystyle{ (z_1, 2) }[/math]. These values are determined by original vector, and the relation between the rotated coordinates and the original ones.
In the original coordinates the two vectors [math]\displaystyle{ x_1 }[/math] and [math]\displaystyle{ x_2 }[/math] are not linearly uncorrelated. In other words, after applying PCA, if there are two principal component, and use those as the new coordinates, then it is guaranteed that the data along the two coordinates have correlation = 0
By rotating the coordinates we performed an orthonormal transform on the data. After the transform, the correlation is removed - which is the whole point of PCA.
As an example of an extreme case where all of the points lie on a straight line, which in the original coordinates system it is needed two coordinates to describe these points.
In short, after rotating the coordinates, we need only one coordinate and clearly the value on the other one is always zero. This example shows PCA's dimension reduction in an extreme case while in real world points may not fit exactly on a line, but only approximately.
PCA applications
As mentioned, PCA is a method to reduce data dimension if possible to principal components such that those PCs cover as much data variation as possible. This technique is useful in different type of applications which involve data with a huge dimension like Data pre-processing, neuroscience, Computer graphics, meteorology, oceanography, gene expression, economics, and finance among of all other applications.
Data usually is represented by lots of variables. In data preprocessing, PCA is a technique to select a subset of variables in order to figure our best model for data. In neuroscience, PCA used to identify the specific properties of a stimulus that increase a neuron’s probability of generating an action potential.
Figures below show some example of PCA in real world applications. Click to enlarge.
-
Finding two principal components of original data in 2D space. Components of data are highly correlated in original space. So they can be substituted by just one principal component, PC1, which has more than %99 of total variance of data.
-
PCA in Neuroscience. Data in different classes in 3 dimensional space is mapped to a 2 dimensional space using PCA. The mapped data is clearly linearly sparable.
Mathematical Details
PCA is a transformation from original space to a linear subspace with a new coordinate system. Each coordinate of this subspace is called a Principle Component. First principal component is the coordinate of this system along which the data points has the maximum variation. That is, if we project the data points along this coordinate, maximum variance of data is obtained (compared to any other vector in original space). Second principal component is the coordinate in the direction of the second greatest variance of the data, and so on.
Lets denote the basis of original space by [math]\displaystyle{ \mathbf{v_1} }[/math], [math]\displaystyle{ \mathbf{v_2} }[/math], ... , [math]\displaystyle{ \mathbf{v_d} }[/math]. Our goal is to find the principal components (coordinate of the linear subspace), denoted by [math]\displaystyle{ \mathbf{u_1} }[/math], [math]\displaystyle{ \mathbf{u_2} }[/math], ... , [math]\displaystyle{ \mathbf{u_p} }[/math] in the hope that [math]\displaystyle{ p \leq d }[/math]. First, we would like to obtain the first principal component [math]\displaystyle{ \mathbf{u_1} }[/math] or the components in the direction of maximum variance. This component can be treated as a vector in the original space and so is written as a linear combination of the basis in original space.
[math]\displaystyle{ \mathbf{u_1}=w_1\mathbf{v_1}+w_2\mathbf{v_2}+...+w_d\mathbf{v_d} }[/math]
Vector [math]\displaystyle{ \mathbf{w} }[/math] contains the weight of each basis in this combination.
[math]\displaystyle{ \mathbf{w}=\begin{bmatrix} w_1\\w_2\\w_3\\...\\w_d \end{bmatrix} }[/math]
Suppose we have n data points in the original space. We represent each data points by [math]\displaystyle{ \mathbf{x_1} }[/math], [math]\displaystyle{ \mathbf{x_2} }[/math], ..., [math]\displaystyle{ \mathbf{x_n} }[/math]. Projection of each point [math]\displaystyle{ \mathbf{x_i} }[/math] on the [math]\displaystyle{ \mathbf{u_1} }[/math] is [math]\displaystyle{ \mathbf{w}^T\mathbf{x_i} }[/math].
Let [math]\displaystyle{ \mathbf{S} }[/math] be the covariance matrix of data points in original space. The variance of the projected data points, denoted by [math]\displaystyle{ \Phi }[/math] is
[math]\displaystyle{ \Phi = Var(\mathbf{w}^T \mathbf{x_i}) = \mathbf{w}^T \mathbf{S} \mathbf{w} }[/math]
we would like to maximize [math]\displaystyle{ \Phi }[/math] over set of all vectors [math]\displaystyle{ \mathbf{w} }[/math] in original space. But, this problem is not yet well-defined, because for any choice of [math]\displaystyle{ \mathbf{w} }[/math], we can increase [math]\displaystyle{ \Phi }[/math] by simply multiplying [math]\displaystyle{ \mathbf{w} }[/math] in a positive scalar greater than one. So, we add the following constraint to the problem to bound the length of vector [math]\displaystyle{ \mathbf{w} }[/math]:
[math]\displaystyle{ max \Phi = \mathbf{w}^T \mathbf{S} \mathbf{w} }[/math]
subject to : [math]\displaystyle{ \mathbf{w}^T \mathbf{w} =1 }[/math]
Using Lagrange Multiplier technique we have:
[math]\displaystyle{ L(\mathbf{w} , \lambda ) = \mathbf{w}^T \mathbf{S} \mathbf{w} - \lambda (\mathbf{w}^T \mathbf{w} - 1 ) }[/math]
By taking derivative of [math]\displaystyle{ L }[/math] w.r.t. primary variable [math]\displaystyle{ \mathbf{w} }[/math] we have:
[math]\displaystyle{ \frac{\partial L}{\partial \mathbf{w}}=( \mathbf{S}^T + \mathbf{S})\mathbf{w} -2\lambda\mathbf{w}= 2\mathbf{S}\mathbf{w} -2\lambda\mathbf{w}= 0 }[/math]
Note that [math]\displaystyle{ \mathbf{S} }[/math] is symmetric so [math]\displaystyle{ \mathbf{S}^T = \mathbf{S} }[/math].
From the above equation we have:
[math]\displaystyle{ \mathbf{S} \mathbf{w} = \lambda\mathbf{w} }[/math].
So [math]\displaystyle{ \mathbf{w} }[/math] is the eigenvector and [math]\displaystyle{ \lambda }[/math] is the eigenvalue of the matrix [math]\displaystyle{ \mathbf{S} }[/math] .(Taking derivative of [math]\displaystyle{ L }[/math] w.r.t. [math]\displaystyle{ \lambda }[/math] just regenerate the constraint of the optimization problem.)
By multiplying both sides of the above equation to [math]\displaystyle{ \mathbf{w}^T }[/math] and considering the constraint, we obtain:
[math]\displaystyle{ \Phi = \mathbf{w}^T \mathbf{S} \mathbf{w} = \mathbf{w}^T \lambda \mathbf{w} = \lambda \mathbf{w}^T \mathbf{w} = \lambda }[/math]
The interesting result is that objective function is equal to eigenvalue of the covariance matrix. So, to obtain the first principle component, which maximizes the objective function, we just need the eigenvector corresponding to the largest eigenvalue of [math]\displaystyle{ \mathbf{S} }[/math]. Subsequently, the second principal component is the eigenvector corresponding to the second largest eigenvalue, and so on.
Principal Component Extraction using Singular Value Decomposition
Singular Value Decomposition (SVD), is a well-known way to decompose any kind of matrix [math]\displaystyle{ \mathbf{A} }[/math] (m by n) into three useful matrices.
[math]\displaystyle{ \mathbf{A} = \mathbf{U}\mathbf{\Sigma}\mathbf{V}^T }[/math].
where [math]\displaystyle{ \mathbf{U} }[/math] is m by m unitary matrix, [math]\displaystyle{ \mathbf{UU^T} = I_m }[/math], each column of [math]\displaystyle{ \mathbf{U} }[/math] is an eigenvector of [math]\displaystyle{ \mathbf{AA^T} }[/math]
[math]\displaystyle{ \mathbf{V} }[/math] is n by n unitary matrix, [math]\displaystyle{ \mathbf{VV^T} = I_n }[/math], each column of [math]\displaystyle{ \mathbf{V} }[/math] is an eigenvector of [math]\displaystyle{ \mathbf{A^TA} }[/math]
[math]\displaystyle{ \mathbf{\Sigma} }[/math] is m by n diagonal matrix, non-zero elements of this matrix are square roots of eigenvalues of [math]\displaystyle{ \mathbf{AA^T} }[/math].
Now, comparing the concepts of PCA and SVD, one may find out that we can perform PCA using SVD.
Lets construct a matrix p by n with our n data points such that each column of this matrix represent one data point in p-dimensional space:
[math]\displaystyle{ \mathbf{X} = [\mathbf{x_1} \mathbf{x_2} .... \mathbf{x_n} ] }[/math]
and make another matrix [math]\displaystyle{ \mathbf{X}^* }[/math] simply by subtracting the mean of data points from [math]\displaystyle{ \mathbf{X} }[/math].
[math]\displaystyle{ \mathbf{X}^* = \mathbf{X} - \mu_X }[/math]
Then we will get a zero-mean version of our data points for which [math]\displaystyle{ \mathbf{X^*} \mathbf{X^{*^T}} }[/math] is the covariance matrix.
So, [math]\displaystyle{ \mathbf{\Sigma} }[/math] and [math]\displaystyle{ \mathbf{U} }[/math] give us the eigenvalues and corresponding eigenvectors of covariance matrix, respectively. We can then easily extract the desired principal components.
MATLAB Example
In this example we use different pictures of a man's face with different facial expressions. But, these pictures are noisy. So the face is not easily recognizable. The data set consists of 1965 pictures each 20 by 28. So dimensionality is 20*28= 560.
Our goal is to remove noise using PCA and of course by means of SVD function in matlab. We know that noise is not the main feature that makes these pictures look different, so noise is among least variance components. We first extract the principal components and then remove those which correspond to the least values of eigenvalues of covariance matrix. Here, eigenvalues are sorted in a descending order from column to column of matrix S. For example, the first column of matrix U, which corresponds to the first element in S, is the first principal component.
We then reconstruct the picture using first d principal components. If d is too large, we can not completely remove the noise. If it is too small, we will loose some information from original data, for example we may loose the facial expression (smile, sadness and etc.). We can change d until we achieve a reasonable balance between noise reduction and information loss.
>> % loading the noisy data, file "noisy" stores our variable X which contains the pictures >> load noisy; >> >> >> % show a sample image in column 1 of matrix X >> imagesc(reshape(X(:,1),20,28)') >> >> % set the color of image to grayscale >> colormap gray >> >> % perform SVD, if X matrix if full rank, will obtain 560 PCs >> [U S V] = svd(X); >> >> d= 10; >> >> % reconstruct X using only the first d principal components and removing noise >> XX = U(:, 1:d)* S(1:d,1:d)*U(:,1:d)' ; >> >> % show image in column 1 of XX which is a noise reduced version >> imagesc(reshape(XX(:,1),20,28)')
PCA Continued, Lagrange Multipliers, Singular Value Decomposition (SVD) (Lecture: Sep. 15, 2014)
Principal Component Analysis (Continued)
PCA is a method to reduce dimensions in data or to extract features from data.
Given a data point in vector form x the goal of PCA is to map x to y where x [math]\displaystyle{ \isin }[/math] [math]\displaystyle{ \real }[/math]d and y [math]\displaystyle{ \isin }[/math] [math]\displaystyle{ \real }[/math]p such that p is much less than d. For example we could map a two-dimensional data point onto any one-dimensional vector in the original 2D space or we could map a three-dimensional data point onto any 2D plane in the original 3D space.
In the case of many data points xi in d-dimensional space:
Xdxn = [x1 x2 ... xn]
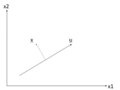
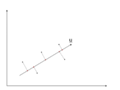
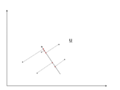
There is any infinite amount of vectors in [math]\displaystyle{ \real }[/math]p to which the points in X can be mapped. When mapping these data points to lower dimensions information will be lost. To preserve as much information as possible the points are mapped to a vector in [math]\displaystyle{ \real }[/math]p which will preserve as much variation in the data as possible. In other words, the data is mapped to the vector in [math]\displaystyle{ \real }[/math]p that will have maximum variance. In the images below, the data points mapped onto u in Mapping 1 have greater variance than in Mapping 2, and thus preserves more information in the data.
-
Two dimensional point mapped onto one dimensional vector
-
Mapping 1: Mapping of a two dimensional data set onto a one dimensional vector
-
Mapping 2: Mapping of a two dimensional data set onto a one dimensional vector
To project n data points onto u compute Y = uTX.
The variance of Y = uTX can be calculated easily as Y is a 1xn vector.
In higher dimensional space [math]\displaystyle{ \real }[/math]q where q > 1 the concept is Covariance.
The variance of projected points in the direction of vector u is given by:
Var(uTX) = uTSu where S is the sample covariance matrix of X.
In finding the first principal component the objective is to find the direction u which will have the greatest variance of the points projected onto it. This can be expressed as the following optimization problem:
maxu uTSu
s.t. uTu = 1
The restriction uTu = 1 is to bound the problem. If the length of u were not restricted, uTSu would go to infinity as the magnitude of u goes to infinity. The selection of 1 as the bound is arbitrary.
Lagrange Multiplier Review
The Lagrange multiplier is a method for solving optimization problems. After constraining the principal component analysis output vector to an arbitrary size the task of maximizing variance in lower dimensional spaces becomes an optimization problem.
Lagrange Multipler Theorem
The method of Lagrange multipliers states that given the optimization problem:
[math]\displaystyle{ max\ f(x,y) }[/math] [math]\displaystyle{ s.t.\ g(x,y) = c }[/math]
The Legrangian is defined by:
[math]\displaystyle{ \ L(x,y,\lambda)= f(x,y) - \lambda(g(x,y)-c) }[/math]
Which implies, where [math]\displaystyle{ \bigtriangledown\ }[/math] is the gradient, that:
[math]\displaystyle{ \bigtriangledown\ f(x,y) = \lambda \bigtriangledown g(x,y) }[/math]
[math]\displaystyle{ \bigtriangledown\ f(x,y) - \lambda \bigtriangledown g(x,y) = 0 }[/math]
In practice,
[math]\displaystyle{ \bigtriangledown\ L }[/math] is calculated by computing the three partial derivatives of [math]\displaystyle{ \ L }[/math] with respect to [math]\displaystyle{ \ x, y, \lambda }[/math], setting them to 0, and solving the resulting system of equations.
That is to say, the solution is given by taking the solution of:
[math]\displaystyle{ \ \bigtriangledown L = \begin{bmatrix} \frac{\partial L}{\partial x}\\\frac{\partial L}{\partial y}\\\frac{\partial L}{\partial \lambda}\end{bmatrix}= 0 }[/math]
which maximize the objective function f(x,y).
Intuitive explanation
The Lagrange multiplier utilizes gradients to find maximum values subject to constraints. A gradient [math]\displaystyle{ \bigtriangledown }[/math] is a vector which collects all of the partial derivatives of a function. Intuitively, this can be thought of as a vector which contains the slopes of all of a graph's tangent lines, or the direction which each point on the graph points.
Finding a point where the gradient of f(x,y) and g(x,y) differ by a factor lambda: [math]\displaystyle{ \bigtriangledown f(x,y) = \lambda \bigtriangledown g(x,y) }[/math] defines a point where our restraint and objective functions are both pointing in the same direction. This is important because if our objective function is pointing in a higher direction than g, increasing our objective function will increase our output, meaning we are not at a maximum. If the restraint is pointing in a higher direction than f, then reducing our input value will yield a higher output. The factor lambda is important because it is adjusted to ensure that the constraint is satisfied and maximized at our current location.
Lagrange Multiplier Example
Objective function: f(x,y) = max x - y
s.t. g(x,y) = x2 + y2 = 1
Calculating the Lagrangian gives:
L(x,y,[math]\displaystyle{ \lambda }[/math]) = x - y - [math]\displaystyle{ \lambda }[/math](x2 + y2 - 1)
Which gives three partial derivatives:
[math]\displaystyle{ \frac{\partial L}{\partial x} = 1 - 2\lambda x = 0 }[/math]
[math]\displaystyle{ \frac{\partial L}{\partial y} = -1 - 2\lambda y = 0 }[/math]
[math]\displaystyle{ \frac{\partial L}{\partial \lambda} = x^2 + y - 1 = 0 }[/math]
This gives two solutions:
Solution 1:
[math]\displaystyle{ x = \frac{\sqrt{2}}{2}, y = \frac{-\sqrt{2}}{2}, \lambda = \frac{1}{\sqrt{2}} }[/math]
Solution 2:
[math]\displaystyle{ x = \frac{-\sqrt{2}}{2}, y = \frac{\sqrt{2}}{2}, \lambda = \frac{1}{\sqrt{2}} }[/math]
Both solutions give extreme values of our objective function. Because this is an optimization problem we are interested in the solution which gives the maximum output.
Singular Value Decomposition Revisited
Any matrix X can be decomposed to the three matrices: [math]\displaystyle{ \ U \Sigma V^T }[/math] where:
[math]\displaystyle{ \ U }[/math] is an m x m matrix of the eigenvector XXT
[math]\displaystyle{ \ \Sigma }[/math] is an m x n matrix containing singular values along the diagonal and zeroes elsewhere
[math]\displaystyle{ \ V^T }[/math] is an orthogonal transpose of a n x n matrix of the eigenvector XTX
Connection of Singular Value Decomposition to PCA
If X is centered (has a mean of 0) then XXT is the covariance matrix.
Using the SVD of X we get:
[math]\displaystyle{ \ XX^T = (U \Sigma V^T)(U \Sigma V^T)^T }[/math]
[math]\displaystyle{ \ XX^T = (U \Sigma V^T)(V \Sigma U^T) }[/math]
[math]\displaystyle{ \ XX^T = U \Sigma V^T V \Sigma U^T }[/math]
[math]\displaystyle{ \ XX^T = U \Sigma I \Sigma U^T }[/math] (By the definition of V)
[math]\displaystyle{ \ XX^T = U \Sigma^2 U^T }[/math]
It is important to remember that U and V are unitary matricies, i.e.: [math]\displaystyle{ U^TU=UU^T=I }[/math] and [math]\displaystyle{ V^TV=VV^T }[/math], each equality using the appropriate dimensional identity matrix..
It is also interesting that the nonsquare matrix formed by taking a subset of the eigenvectors contained in [math]\displaystyle{ U }[/math] or [math]\displaystyle{ V }[/math] has the property that [math]\displaystyle{ V^TV=I }[/math] (the appropriate lower dimensional identity matrix)
Matlab Example, Dimensionality Reduction Using PCA
The following example shows how principal component analysis can be used to identify between a handwritten 2 and a handwritten 3. In the example the original images are each 64 pixels and thus 64 dimensional. By reducing the dimensionality to 2D the 2's and 3's are visualized on a 2D plot and can it could be estimated as to which digit one was.
>> % load file containing 200 handwritten 2's and 200 handwritten 3's >> load 2_3; >> >> who? >> % displays that X is 64x400 matrix containing the handwritten digits >> >> imagesc(reshape(X(:,1),8,8)') >> colormap gray >> % displays first image, a handwritten 2 >> >> M = mean(X,2)*ones(1,400); >> Xb = X-M >> % M is matrix of mean of columns repeated 400 times >> % Xb is data centred about mean >> >> % Now use SVD to find eigenvectors, eigenvalues of Xb >> [U S V] = svd(Xb); >> >> % 2D projection on points >> Y = U(:,1:2)'*X; >> >> % 2D projections of 2's >> plot(Y(1,1:200),Y(2,1:200),'.') >> >> % 2D projections of 3's >> plot(Y(1,201:400),Y(2,201:400),'.')
PCA Algorithm, Dual PCA, and Kernel PCA (Lecture: Sep. 17, 2014)
PCA Algorithm (Algorithm 1)
Recover Basis:
To find the basis for PCA we first remove the mean of the sample data and calculate
- [math]\displaystyle{ XX^T = \sum_{i=1}^n x_i x_i^T }[/math]
Then let [math]\displaystyle{ U }[/math] be the eigenvectors of [math]\displaystyle{ XX^T }[/math] corresponding to the top [math]\displaystyle{ \,d }[/math] eigenvalues.
Encode Training Data:
To encode our data using PCA we let
- [math]\displaystyle{ \,Y = U^TX }[/math]
where [math]\displaystyle{ Y }[/math] is an encoded matrix of the original data
Reconstruct Training Data:
To project our data back to the higher dimension
- [math]\displaystyle{ \hat{X} = UY = UU^T X }[/math]
Encode Test Example:
- [math]\displaystyle{ \,y = U^T x }[/math]
where [math]\displaystyle{ \,y }[/math] is an encoding of [math]\displaystyle{ \,x }[/math]
Reconstruct Test Example:
- [math]\displaystyle{ \hat{x} = Uy = UU^T x }[/math]
Dual Principal Component Analysis
Motivation
In some cases the dimension of our data can be much larger than the number of data points [math]\displaystyle{ ( d \gt \gt n ) }[/math]. For example, suppose we are interested in weekly closing rates of the Dow Jones Industrial Average, S&P 500 Index, and the NASDAQ Composite Index over the past forty years. In this example we only have three data points [math]\displaystyle{ ( n = 3 ) }[/math] while our dimension is over two thousand! [math]\displaystyle{ ( d \approx 2080 ) }[/math] This means that the matrix [math]\displaystyle{ XX^T }[/math] used to find the basis [math]\displaystyle{ U }[/math] is [math]\displaystyle{ 2080\times2080 }[/math]. Another example is that if we want to apply PCA to the genes of some patients, the number of genes studied could be way larger than the number of patients. In these cases, a less compute-intensive way of recovering the basis is desired.
In PCA recall that we used the Singular Value Decomposition:
- [math]\displaystyle{ \,X = U \Sigma V^T }[/math]
Multiply both sides by V to get:
- [math]\displaystyle{ \,X V = U \Sigma V^T V }[/math]
Since [math]\displaystyle{ V^T V = I }[/math] (the identity matrix) the SVD can be rearranged as
- [math]\displaystyle{ \,X V = U \Sigma }[/math]
Then, multiply both sides by [math]\displaystyle{ \Sigma^-1 }[/math] to get:
- [math]\displaystyle{ \,U = X V \Sigma^-1 }[/math]
And so [math]\displaystyle{ U }[/math] can be calculated in terms of [math]\displaystyle{ V }[/math] and [math]\displaystyle{ \Sigma }[/math]. [math]\displaystyle{ V }[/math] is the eigenvectors of [math]\displaystyle{ X^T X_{n \times n} }[/math], so it is much easier to find than [math]\displaystyle{ U }[/math] because [math]\displaystyle{ n\lt \lt d }[/math].
Algorithm
We can replace all instances of [math]\displaystyle{ \,U }[/math] in Algorithm 1 with [math]\displaystyle{ \,U = X V \Sigma^{-1} }[/math] to obtain the algorithm for Dual PCA.
Recover Basis:
For the basis of Dual PCA, we calculate
- [math]\displaystyle{ XX^T }[/math]
then let [math]\displaystyle{ V }[/math] be the eigenvectors of [math]\displaystyle{ XX^T }[/math] with respect to the top [math]\displaystyle{ d }[/math] eigenvalues. Finally we let [math]\displaystyle{ \Sigma }[/math] be the diagonal matrix of square roots of the top [math]\displaystyle{ d }[/math] eigenvalues.
Encode Training Data:
Using Dual PCA to encode the data, let
- [math]\displaystyle{ \,Y = U^TX = \Sigma V^T }[/math]
where [math]\displaystyle{ Y }[/math] is an encoded matrix of the original data
Reconstruct Training Data:
Project the data back to the higher dimension by
- [math]\displaystyle{ \hat{X} = UY = U \Sigma V^T = X V \Sigma^{-1} \Sigma V^T = X V V^T }[/math]
Encode Test Example:
- [math]\displaystyle{ \,y = U^T x = \Sigma^{-1} V^T X^T x = \Sigma^{-1} V^T X^T x }[/math]
where [math]\displaystyle{ \,y }[/math] is an encoding of [math]\displaystyle{ \,x }[/math]
Reconstruct Test Example:
- [math]\displaystyle{ \hat{x} = Uy = UU^T x = X V \Sigma^{-2} V^T X^T x = X V \Sigma^{-2} V^T X^T x }[/math]
Note that the steps of Reconstructing training data and Reconstructioning test example still depend on [math]\displaystyle{ d }[/math], and therefore still will be impractical in the case that the original dimensionality of the data ([math]\displaystyle{ d }[/math]) is very large
Kernel Principle Component Analysis
Kernel Methods
Kernel methods are a class of algorithms used for analyzing patterns and measuring similarity. They are used for finding relations within a data set. Kernel methods map data into a higher-dimensional space to make it easier to find these relations. While PCA is designed for linear variabilities, a lot of higher-dimensional data are nonlinear. This is where Kernel PCA can be useful to help model the data variability when it comes to a nonlinear manifold.
Some common examples of kernels include:
1. Linear Kernel:
- [math]\displaystyle{ k_{ij} = \lt x_{i},x_{j}\gt }[/math]
2. Polynomial Kernel:
- [math]\displaystyle{ k_{ij} = (1+\lt x_{i},x_{j}\gt )^P }[/math]
3. Gaussian Kernel:
- [math]\displaystyle{ k_{ij} = exp(||x_{i}-x_{j}||^2/2\sigma^2) }[/math]
Other kernel examples can be found here:
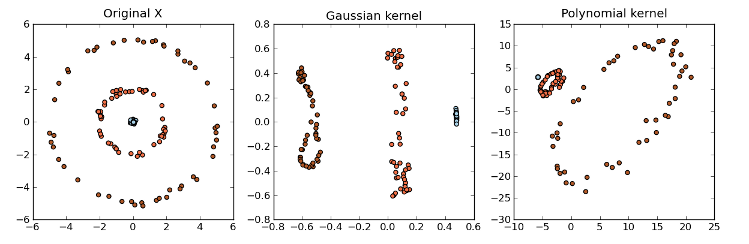
Kernel PCA
With the Kernel PCA, we take the original or observed space and map it to the feature space and then map the feature space to the embedded space of a lower dimension.
- [math]\displaystyle{ X_{observed space} -\gt \Eta_{feature space} -\gt Y_{embedded space} }[/math]
In the Kernel PCA algorithm, we can use the same argument as PCA and again use the Standard Value Decomposition (SVD) where:
- [math]\displaystyle{ \Phi(X) = U \Sigma V^T }[/math]
where [math]\displaystyle{ U }[/math] contains the eigenvectors of [math]\displaystyle{ \Phi(X)\Phi(X)^T }[/math]
The algorithm is similar to the Dual PCA algorithm except that the training and the test data cannot be reconstructed since [math]\displaystyle{ \Phi(X) }[/math] is unknown.
Algorithm
Recover Basis:
For the basis of Kernel PCA, we calculate
- [math]\displaystyle{ K = \Phi(X)^T \Phi(X) }[/math]
using the kernel [math]\displaystyle{ K }[/math] and let [math]\displaystyle{ V }[/math] be the eigenvectors of [math]\displaystyle{ \Phi(X)^T \Phi(X) }[/math] with respect to the top [math]\displaystyle{ p }[/math] eigenvalues. Finally we let [math]\displaystyle{ \Sigma }[/math] be the diagonal matrix of square roots of the top [math]\displaystyle{ p }[/math] eigenvalues.
Encode Training Data:
Using Kernel PCA to encode the data, let
- [math]\displaystyle{ \,Y = U^T \Phi(X) = \Sigma V^T }[/math]
Recall [math]\displaystyle{ U = \Phi(X) V \Sigma^{-1} }[/math]:
- [math]\displaystyle{ \,Y = U^T \Phi(X) = \Sigma^{-1} V^T \Phi(X)^T \Phi(X) = \Sigma^{-1} V^T K(X,X) }[/math]
where [math]\displaystyle{ Y }[/math] is an encoded matrix of the image of the original data in the function [math]\displaystyle{ \Phi }[/math]. [math]\displaystyle{ Y }[/math] is computable without knowledge of [math]\displaystyle{ \Phi(X) }[/math] via the kernel function.
Reconstruct Training Data:
- [math]\displaystyle{ \hat{\Phi(X)} = UY = UU^T\Phi(X)=\Phi(X)V\Sigma^{-1}\Sigma V^T=\Phi(X) V V^T }[/math]
- [math]\displaystyle{ \hat{X} = \Phi^{-1}(\hat{\Phi(X)}) }[/math]
However, [math]\displaystyle{ \Phi(X) }[/math] is unknown so we cannot reconstruct the data.
Encode Test Example:
- [math]\displaystyle{ \,y = U^T \Phi(x) = \Sigma^{-1} V^T \Phi(X)^T \Phi(x) = \Sigma^{-1} V^T K(X,x) }[/math]
This is possible because we know that [math]\displaystyle{ \Phi(X)^T \Phi(X) = K(X,x) }[/math].
Recall
[math]\displaystyle{ U = \Phi(X) V \Sigma^{-1} }[/math]:
[math]\displaystyle{ \Phi(X) }[/math] is the transformation of the training data [math]\displaystyle{ X_{d \times n} }[/math] while [math]\displaystyle{ \Phi(x) }[/math] is the transformation of the test example [math]\displaystyle{ x_{d \times 1} }[/math]
Reconstruct Test Example:
- [math]\displaystyle{ \hat{\Phi(x)} = Uy = UU^T \Phi(x) = \Phi(X) V \Sigma^{-2} V^T \Phi(X)^T \Phi(x) = \Phi(X) V \Sigma^{-2} V^T K(X,x) }[/math]
Once again, since [math]\displaystyle{ \Phi(X) }[/math] is unknown, we cannot reconstruct the data.
Lecture 3 Centering for Kernel PCA and Multidimensional Scaling
Centering for Kernel PCA
In the derivation of Kernel PCA we assumed that [math]\displaystyle{ \sum_{i=1}^n\Phi(x_i)=0 }[/math] which is improbable for a random data set and an arbitrary mapping [math]\displaystyle{ \Phi }[/math]. We must be able to ensure this condition though we don't know [math]\displaystyle{ \Phi }[/math].
Let [math]\displaystyle{ \mu=\frac{1}{n}\sum_{j=1}^n\Phi(x_j) }[/math]
Define the centered transformation (not computable)
- [math]\displaystyle{ \tilde{\Phi}:x\mapsto(\Phi(x)-\mu) }[/math]
and the centered kernel (computable)
- [math]\displaystyle{ \tilde{K}:(x,y)\mapsto \tilde{\Phi}(x)^T \tilde{\Phi}(y) }[/math]
Then
- [math]\displaystyle{ \tilde{K}(x,y) = (\Phi(x)-\mu)^T (\Phi(y)-\mu)=\Phi(x)^T\Phi(y)-\mu^T\Phi(x)-\mu^T\Phi(y)+\mu^T\mu }[/math]
[math]\displaystyle{ =K(x,y)-\frac{1}{n}\sum_{j=1}^n\Phi(x_j)^T\Phi(x)-\frac{1}{n}\sum_{j=1}^n\Phi(x_j)^T\Phi(y)+\frac{1}{n^2}\sum_{j=1}^n\sum_{i=1}^n\Phi(x_j)^T\Phi(x_i) }[/math]
[math]\displaystyle{ =K(x,y)-\frac{1}{n}\sum_{j=1}^n K(x_j,x)-\frac{1}{n}\sum_{j=1}^n K(x_j,y)+\frac{1}{n^2}\sum_{j=1}^n\sum_{i=1}^n K(x_j,x_i) }[/math]
[math]\displaystyle{ =K(x,y)-\frac{1}{n}\bold{1}^TK(X,x)-\frac{1}{n}\bold{1}^TK(X,y)+\frac{1}{n^2}\bold{1}^TK(X,X)\bold{1} }[/math]
where [math]\displaystyle{ \bold{1}=([1,1,...,1]_{1\times n})^T }[/math]
Here [math]\displaystyle{ X_{p\times n} }[/math] and [math]\displaystyle{ \{x_j\}_{j=1}^n }[/math] denote the data set matrix and elements respectively and [math]\displaystyle{ x,y }[/math] represent test elements at which the centered kernel is evaluated.
Multidimensional Scaling
Multidimensional Scaling (MDS) is a dimension reduction method that attempts to preserve the pair-wise distance between data points. For example, Suppose we want to map a set of points in X where each element [math]\displaystyle{ x \in \mathbb{R}^d }[/math] is mapped to [math]\displaystyle{ y \in \mathbb{R}^p }[/math]. We want to preserve the pair-wise distances such that [math]\displaystyle{ d(y_i, y_j) }[/math] is relatively the same as [math]\displaystyle{ d(y_i, y_j) }[/math]
MDS Example
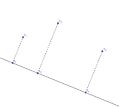
We can map points in 2 dimensions to points in 1 dimension by projecting the points on to a straight line:
-
Multidimensional scaling from 2D to 1D
Note that the pair-wise distances are relatively preserved. However, the triangle inequality is {\bf not} preserved: [math]\displaystyle{ d(x_1,x_2)+d(x_2,x_3) \gt d(x_1, x_3) }[/math] But [math]\displaystyle{ d(y_1,y_2)+d(y_2,y_3) }[/math] is not greater than [math]\displaystyle{ d(y_1, y_3) }[/math]
Distance Matrix
Given a [math]\displaystyle{ d x n }[/math] matrix [math]\displaystyle{ X }[/math] The distance matrix is defined by:
- [math]\displaystyle{ D^X_{n*n} = \begin{bmatrix} d_11\\d_12\\w_3\\...\\w_d \end{bmatrix} }[/math]
Theorem
If [math]\displaystyle{ D^{(X)} }[/math] is a distance matrix and if [math]\displaystyle{ K=-\frac{1}{2}HD^{(X)}H }[/math]
Then [math]\displaystyle{ D^{(X)} }[/math] is Euclidean iff [math]\displaystyle{ K }[/math] is Positive Semi-Definite
Where [math]\displaystyle{ H=I-\frac{1}{n}\bold{1}^T\bold{1} }[/math] and [math]\displaystyle{ \bold{1}=([1,1,...,1]_{1\times n})^T }[/math]