stat841f10
Proposal Fall 2010
Editor sign up
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: Provide a summary for each topic here.. Please improve this article if you can. (October 8 2010) | small = | smallimage = | smallimageright = | smalltext = }}
Digest
Reference Textbook
The Elements of Statistical Learning: Data Mining, Inference, and Prediction, Second Edition, February 2009 Trevor Hastie, Robert Tibshirani, Jerome Friedman (3rd Edition is available)
Classification - September 21, 2010
Classification
Statistical classification, or simply known as classification, is an area of supervised learning that addresses the problem of how to systematically assign unlabeled (classes unknown) novel data to their labels (classes or groups or types) by using knowledge of their features (characteristics or attributes) that are obtained from observation and/or measurement. A classifier is a specific technique or method for performing classification. To classify new data, a classifier first uses labeled (classes are known) training data to train a model, and then it uses a function known as its classification rule to assign a label to each new data input after feeding the input's known feature values into the model to determine how much the input belongs to each class.
Classification has been an important task for people and society since the beginnings of history. According to this link, the earliest application of classification in human society was probably done by prehistory peoples for recognizing which wild animals were beneficial to people and which ones were harmful, and the earliest systematic use of classification was done by the famous Greek philosopher Aristotle (384 BC - 322 BC) when he, for example, grouped all living things into the two groups of plants and animals. Classification is generally regarded as one of four major areas of statistics, with the other three major areas being regression, clustering, and dimensionality reduction (feature extraction or manifold learning). Please be noted that some people consider classification to be a broad area that consists of both supervised and unsupervised methods of classifying data. In this view, as can be seen in this link, clustering is simply a special case of classification and it may be called unsupervised classification.
In classical statistics, classification techniques were developed to learn useful information using small data sets where there is usually not enough of data. When machine learning was developed after the application of computers to statistics, classification techniques were developed to work with very large data sets where there is usually too many data. A major challenge facing data mining using machine learning is how to efficiently find useful patterns in very large amounts of data. An interesting quote that describes this problem quite well is the following one made by the retired Yale University Librarian Rutherford D. Rogers, a link to a source of which can be found here.
"We are drowning in information and starving for knowledge."
- Rutherford D. Rogers
In the Information Age, machine learning when it is combined with efficient classification techniques can be very useful for data mining using very large data sets. This is most useful when the structure of the data is not well understood but the data nevertheless exhibit strong statistical regularity. Areas in which machine learning and classification have been successfully used together include search and recommendation (e.g. Google, Amazon), automatic speech recognition and speaker verification, medical diagnosis, analysis of gene expression, drug discovery etc.
The formal mathematical definition of classification is as follows:
Definition: Classification is the prediction of a discrete random variable [math]\displaystyle{ \mathcal{Y} }[/math] from another random variable [math]\displaystyle{ \mathcal{X} }[/math], where [math]\displaystyle{ \mathcal{Y} }[/math] represents the label assigned to a new data input and [math]\displaystyle{ \mathcal{X} }[/math] represents the known feature values of the input.
A set of training data used by a classifier to train its model consists of [math]\displaystyle{ \,n }[/math] independently and identically distributed (i.i.d) ordered pairs [math]\displaystyle{ \,\{(X_{1},Y_{1}), (X_{2},Y_{2}), \dots , (X_{n},Y_{n})\} }[/math], where the values of the [math]\displaystyle{ \,ith }[/math] training input's feature values [math]\displaystyle{ \,X_{i} = (\,X_{i1}, \dots , X_{id}) \in \mathcal{X} \subset \mathbb{R}^{d} }[/math] is a d-dimensional vector and the label of the [math]\displaystyle{ \, ith }[/math] training input is [math]\displaystyle{ \,Y_{i} \in \mathcal{Y} }[/math] that can take a finite number of values. The classification rule used by a classifier has the form [math]\displaystyle{ \,h: \mathcal{X} \mapsto \mathcal{Y} }[/math]. After the model is trained, each new data input whose feature values is [math]\displaystyle{ \,x }[/math] is given the label [math]\displaystyle{ \,\hat{Y}=h(x) }[/math].
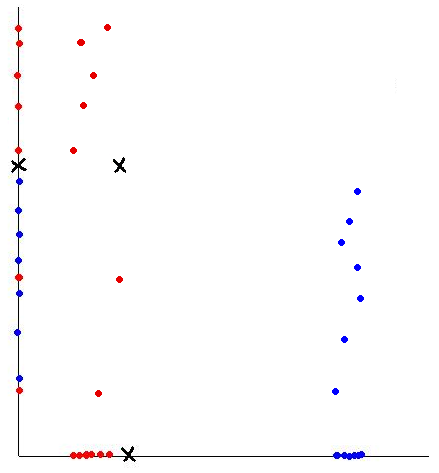
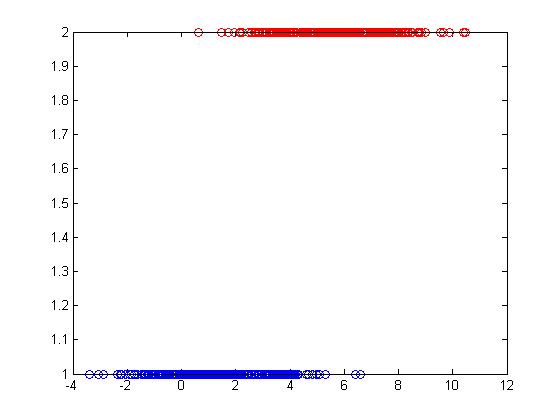
As an example, if we would like to classify some vegetables and fruits, then our training data might look something like the one shown in the following picture from Professor Ali Ghodsi's Fall 2010 STAT 841 slides.
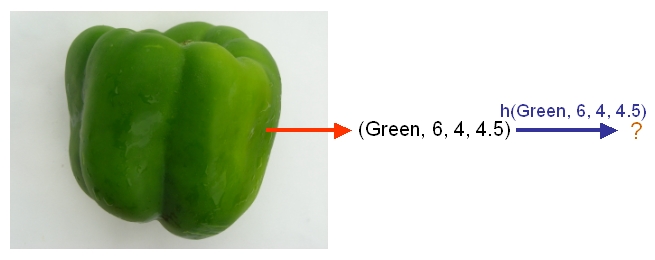
After we have selected a classifier and then built our model using our training data, we could use the classifier's classification rule [math]\displaystyle{ \ h }[/math] to classify any newly-given vegetable or fruit such as the one shown in the following picture from Professor Ali Ghodsi's Fall 2010 STAT 841 slides after first obtaining its feature values.
As another example, suppose we wish to classify newly-given fruits into apples and oranges by considering three features of a fruit that comprise its color, its diameter, and its weight. After selecting a classifier and constructing a model using training data [math]\displaystyle{ \,\{(X_{color, 1}, X_{diameter, 1}, X_{weight, 1}, Y_{1}), \dots , (X_{color, n}, X_{diameter, n}, X_{weight, n}, Y_{n})\} }[/math], we could then use the classifier's classification rule [math]\displaystyle{ \,h }[/math] to assign any newly-given fruit having known feature values [math]\displaystyle{ \,x = (\,x_{color}, x_{diameter} , x_{weight}) }[/math] the label [math]\displaystyle{ \, \hat{Y}=h(x) \in \mathcal{Y}= \{apple,orange\} }[/math].
Error rate
The empirical error rate (or training error rate) of a classifier having classification rule [math]\displaystyle{ \,h }[/math] is defined as the frequency at which [math]\displaystyle{ \,h }[/math] does not correctly classify the data inputs in the training set, i.e., it is defined as [math]\displaystyle{ \,\hat{L}_{n} = \frac{1}{n} \sum_{i=1}^{n} I(h(X_{i}) \neq Y_{i}) }[/math], where [math]\displaystyle{ \,I }[/math] is an indicator variable and [math]\displaystyle{ \,I = \left\{\begin{matrix} 1 &\text{if } h(X_i) \neq Y_i \\ 0 &\text{if } h(X_i) = Y_i \end{matrix}\right. }[/math]. Here, [math]\displaystyle{ \,X_{i} \in \mathcal{X} }[/math] and [math]\displaystyle{ \,Y_{i} \in \mathcal{Y} }[/math] are the known feature values and the true class of the [math]\displaystyle{ \,ith }[/math] training input, respectively.
The true error rate [math]\displaystyle{ \,L(h) }[/math] of a classifier having classification rule [math]\displaystyle{ \,h }[/math] is defined as the probability that [math]\displaystyle{ \,h }[/math] does not correctly classify any new data input, i.e., it is defined as [math]\displaystyle{ \,L(h)=P(h(X) \neq Y) }[/math]. Here, [math]\displaystyle{ \,X \in \mathcal{X} }[/math] and [math]\displaystyle{ \,Y \in \mathcal{Y} }[/math] are the known feature values and the true class of that input, respectively.
In practice, the empirical error rate is obtained to estimate the true error rate, whose value is impossible to be known because the parameter values of the underlying process cannot be known but can only be estimated using available data. The empirical error rate, in practice, estimates the true error rate quite well in that, as mentioned here, it is an unbiased estimator of the true error rate.
Bayes Classifier
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: In response to the previous tag: The naive Bayes classifier is a special (simpler) case of the Bayes classifier. It uses an extra assumption: that the presence (or absence) of a particular feature of a class is unrelated to the presence (or absence) of any other feature. This assumption allows for an easier likelihood function [math]\displaystyle{ \,f_y(x) }[/math] in the equation:
- [math]\displaystyle{ \begin{align} P(Y=y|X=x) &=\frac{f_y(x)\pi_y}{\Sigma_{\forall i \in \mathcal{Y}} f_i(x)\pi_i} \end{align} }[/math]
The simper form of the likelihood function seen in the naive Bayes is:
- [math]\displaystyle{ \begin{align} f_y(x) = P(X=x|Y=y) = {\prod_{i=1}^{n} P(X_{i}=x_{i}|Y=y)} \end{align} }[/math]
The Bayes classifier taught in class was not the naive Bayes classifier. Perhaps a comment should be made about the naive Bayes classifier in the body of the text. Please improve this article if you can. (October 14 2010) | small = | smallimage = | smallimageright = | smalltext = }}
A Bayes classifier is a simple probabilistic classifier based on applying Bayes' Theorem (from Bayesian statistics) with strong (naive) independence assumptions. A more descriptive term for the underlying probability model would be "independent feature model".
In simple terms, a Bayes classifier assumes that the presence (or absence) of a particular feature of a class is unrelated to the presence (or absence) of any other feature. For example, a fruit may be considered to be an apple if it is red, round, and about 4" in diameter. Even if these features depend on each other or upon the existence of the other features, a Bayes classifier considers all of these properties to independently contribute to the probability that this fruit is an apple.
Depending on the precise nature of the probability model, naive Bayes classifiers can be trained very efficiently in a supervised learning setting. In many practical applications, parameter estimation for Bayes models uses the method of maximum likelihood; in other words, one can work with the naive Bayes model without believing in Bayesian probability or using any Bayesian methods.
In spite of their design and apparently over-simplified assumptions, naive Bayes classifiers have worked quite well in many complex real-world situations. In 2004, analysis of the Bayesian classification problem has shown that there are some theoretical reasons for the apparently unreasonable efficacy of Bayes classifiers [1]. Still, a comprehensive comparison with other classification methods in 2006 showed that Bayes classification is outperformed by more current approaches, such as boosted trees or random forests[2].
An advantage of the naive Bayes classifier is that it requires a small amount of training data to estimate the parameters (means and variances of the variables) necessary for classification. Because independent variables are assumed, only the variances of the variables for each class need to be determined and not the entire covariance matrix.
After training its model using training data, the Bayes classifier classifies any new data input in two steps. First, it uses the input's known feature values and the Bayes formula to calculate the input's posterior probability of belonging to each class. Then, it uses its classification rule to place the input into the most-probable class, which is the one associated with the input's largest posterior probability.
In mathematical terms, for a new data input having feature values [math]\displaystyle{ \,(X = x)\in \mathcal{X} }[/math], the Bayes classifier labels the input as [math]\displaystyle{ (Y = y) \in \mathcal{Y} }[/math], such that the input's posterior probability [math]\displaystyle{ \,P(Y = y|X = x) }[/math] is maximum over all of the members of [math]\displaystyle{ \mathcal{Y} }[/math].
Suppose there are [math]\displaystyle{ \,k }[/math] classes and we are given a new data input having feature values [math]\displaystyle{ \,x }[/math]. The following derivation shows how the Bayes classifier finds the input's posterior probability [math]\displaystyle{ \,P(Y = y|X = x) }[/math] of belonging to each class [math]\displaystyle{ y \in \mathcal{Y} }[/math].
- [math]\displaystyle{ \begin{align} P(Y=y|X=x) &= \frac{P(X=x|Y=y)P(Y=y)}{P(X=x)} \\ &=\frac{P(X=x|Y=y)P(Y=y)}{\Sigma_{\forall i \in \mathcal{Y}}P(X=x|Y=i)P(Y=i)} \end{align} }[/math]
Here, [math]\displaystyle{ \,P(Y=y|X=x) }[/math] is known as the posterior probability as mentioned above, [math]\displaystyle{ \,P(Y=y) }[/math] is known as the prior probability, [math]\displaystyle{ \,P(X=x|Y=y) }[/math] is known as the likelihood, and [math]\displaystyle{ \,P(X=x) }[/math] is known as the evidence.
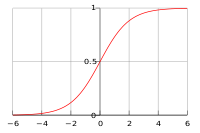
In the special case where there are two classes, i.e., [math]\displaystyle{ \, \mathcal{Y}=\{0, 1\} }[/math], the Bayes classifier makes use of the function [math]\displaystyle{ \,r(x)=P\{Y=1|X=x\} }[/math] which is the posterior probability of a new data input having feature values [math]\displaystyle{ \,x }[/math] belonging to the class [math]\displaystyle{ \,Y = 1 }[/math]. Following the above derivation for the posterior probabilities of a new data input, the Bayes classifier calculates [math]\displaystyle{ \,r(x) }[/math] as follows:
- [math]\displaystyle{ \begin{align} r(x)&=P(Y=1|X=x) \\ &=\frac{P(X=x|Y=1)P(Y=1)}{P(X=x)}\\ &=\frac{P(X=x|Y=1)P(Y=1)}{P(X=x|Y=1)P(Y=1)+P(X=x|Y=0)P(Y=0)} \end{align} }[/math]
The Bayes classifier's classification rule [math]\displaystyle{ \,h^*: \mathcal{X} \mapsto \mathcal{Y} }[/math], then, is
- [math]\displaystyle{ \, h^*(x)= \left\{\begin{matrix} 1 &\text{if } \hat r(x)\gt \frac{1}{2} \\ 0 &\mathrm{otherwise} \end{matrix}\right. }[/math].
Here, [math]\displaystyle{ \,x }[/math] is the feature values of a new data input and [math]\displaystyle{ \hat r(x) }[/math] is the estimated value of the function [math]\displaystyle{ \,r(x) }[/math] given by the Bayes classifier's model after feeding [math]\displaystyle{ \,x }[/math] into the model. Still in this special case of two classes, the Bayes classifier's decision boundary is defined as the set [math]\displaystyle{ \,D(h)=\{x: P(Y=1|X=x)=P(Y=0|X=x)\} }[/math]. The decision boundary [math]\displaystyle{ \,D(h) }[/math] essentially combines together the trained model and the decision function [math]\displaystyle{ \,h^* }[/math], and it is used by the Bayes classifier to assign any new data input to a label of either [math]\displaystyle{ \,Y = 0 }[/math] or [math]\displaystyle{ \,Y = 1 }[/math] depending on which side of the decision boundary the input lies in. From this decision boundary, it is easy to see that, in the case where there are two classes, the Bayes classifier's classification rule can be re-expressed as
- [math]\displaystyle{ \, h^*(x)= \left\{\begin{matrix} 1 &\text{if } P(Y=1|X=x)\gt P(Y=0|X=x) \\ 0 &\mathrm{otherwise} \end{matrix}\right. }[/math].
Bayes Classification Rule Optimality Theorem The Bayes classifier is the optimal classifier in that it results in the least possible true probability of misclassification for any given new data input, i.e., for any generic classifier having classification rule [math]\displaystyle{ \,h }[/math], it is always true that [math]\displaystyle{ \,L(h^*(x)) \le L(h(x)) }[/math]. Here, [math]\displaystyle{ \,L }[/math] represents the true error rate, [math]\displaystyle{ \,h^* }[/math] is the Bayes classifier's classification rule, and [math]\displaystyle{ \,x }[/math] is any given data input's feature values.
Although the Bayes classifier is optimal in the theoretical sense, other classifiers may nevertheless outperform it in practice. The reason for this is that various components which make up the Bayes classifier's model, such as the likelihood and prior probabilities, must either be estimated using training data or be guessed with a certain degree of belief. As a result, the estimated values of the components in the trained model may deviate quite a bit from their true population values, and this can ultimately cause the calculated posterior probabilities of inputs to deviate quite a bit from their true values. Estimation of all these probability functions, as likelihood, prior probability, and evidence function is a very expensive task, computationally, which also makes some other classifiers more favorable than Bayes classifier.
A detailed proof of this theorem is available here.
Defining the classification rule:
In the special case of two classes, the Bayes classifier can use three main approaches to define its classification rule [math]\displaystyle{ \,h^* }[/math]:
- 1) Empirical Risk Minimization: Choose a set of classifiers [math]\displaystyle{ \mathcal{H} }[/math] and find [math]\displaystyle{ \,h^*\in \mathcal{H} }[/math] that minimizes some estimate of the true error rate [math]\displaystyle{ \,L(h^*) }[/math].
- 2) Regression: Find an estimate [math]\displaystyle{ \hat r }[/math] of the function [math]\displaystyle{ x }[/math] and define
- [math]\displaystyle{ \, h^*(x)= \left\{\begin{matrix} 1 &\text{if } \hat r(x)\gt \frac{1}{2} \\ 0 &\mathrm{otherwise} \end{matrix}\right. }[/math].
- 3) Density Estimation: Estimate [math]\displaystyle{ \,P(X=x|Y=0) }[/math] from the [math]\displaystyle{ \,X_{i} }[/math]'s for which [math]\displaystyle{ \,Y_{i} = 0 }[/math], estimate [math]\displaystyle{ \,P(X=x|Y=1) }[/math] from the [math]\displaystyle{ \,X_{i} }[/math]'s for which [math]\displaystyle{ \,Y_{i} = 1 }[/math], and estimate [math]\displaystyle{ \,P(Y = 1) }[/math] as [math]\displaystyle{ \,\frac{1}{n} \sum_{i=1}^{n} Y_{i} }[/math]. Then, calculate [math]\displaystyle{ \,\hat r(x) = \hat P(Y=1|X=x) }[/math] and define
- [math]\displaystyle{ \, h^*(x)= \left\{\begin{matrix} 1 &\text{if } \hat r(x)\gt \frac{1}{2} \\ 0 &\mathrm{otherwise} \end{matrix}\right. }[/math].
Typically, the Bayes classifier uses approach 3 to define its classification rule. These three approaches can easily be generalized to the case where the number of classes exceeds two.
Multi-class classification:
Suppose there are [math]\displaystyle{ \,k }[/math] classes, where [math]\displaystyle{ \,k \ge 2 }[/math].
In the above discussion, we introduced the Bayes formula for this general case:
- [math]\displaystyle{ \begin{align} P(Y=y|X=x) &=\frac{P(X=x|Y=y)P(Y=y)}{\Sigma_{\forall i \in \mathcal{Y}}P(X=x|Y=i)P(Y=i)} \end{align} }[/math]
which can re-worded as:
- [math]\displaystyle{ \begin{align} P(Y=y|X=x) &=\frac{f_y(x)\pi_y}{\Sigma_{\forall i \in \mathcal{Y}} f_i(x)\pi_i} \end{align} }[/math]
Here, [math]\displaystyle{ \,f_y(x) = P(X=x|Y=y) }[/math] is known as the likelihood function and [math]\displaystyle{ \,\pi_y = P(Y=y) }[/math] is known as the prior probability.
In the general case where there are at least two classes, the Bayes classifier uses the following theorem to assign any new data input having feature values [math]\displaystyle{ \,x }[/math] into one of the [math]\displaystyle{ \,k }[/math] classes.
Theorem
- Suppose that [math]\displaystyle{ \mathcal{Y}= \{1, \dots, k\} }[/math], where [math]\displaystyle{ \,k \ge 2 }[/math]. Then, the optimal classification rule is [math]\displaystyle{ \,h^*(x) = arg max_{i} P(Y=i|X=x) }[/math], where [math]\displaystyle{ \,i \in \{1, \dots, k\} }[/math].
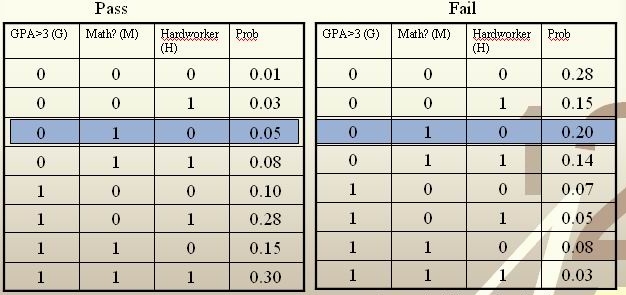
Example: We are going to predict if a particular student will pass STAT 441/841. There are two classes represented by [math]\displaystyle{ \, \mathcal{Y}\in \{ 0,1 \} }[/math], where 1 refers to pass and 0 refers to fail. Suppose that the prior probabilities are estimated or guessed to be [math]\displaystyle{ \,\hat P(Y = 1) = \hat P(Y = 0) = 0.5 }[/math]. We have data on past student performances, which we shall use to train the model. For each student, we know the following:
- Whether or not the student’s GPA was greater than 3.0 (G).
- Whether or not the student had a strong math background (M).
- Whether or not the student was a hard worker (H).
- Whether or not the student passed or failed the course. Note: these are the known y values in the training data.
These known data are summarized in the following tables:
For each student, his/her feature values is [math]\displaystyle{ \, x = \{G, M, H\} }[/math] and his or her class is [math]\displaystyle{ \, y \in \{0, 1\} }[/math].
Suppose there is a new student having feature values [math]\displaystyle{ \, x = \{0, 1, 0\} }[/math], and we would like to predict whether he/she would pass the course. [math]\displaystyle{ \,\hat r(x) }[/math] is found as follows:
[math]\displaystyle{ \, \hat r(x) = P(Y=1|X =(0,1,0))=\frac{P(X=(0,1,0)|Y=1)P(Y=1)}{P(X=(0,1,0)|Y=0)P(Y=0)+P(X=(0,1,0)|Y=1)P(Y=1)}=\frac{0.05*0.5}{0.05*0.5+0.2*0.5}=\frac{0.025}{0.075}=\frac{1}{3}\lt \frac{1}{2}. }[/math]
The Bayes classifier assigns the new student into the class [math]\displaystyle{ \, h^*(x)=0 }[/math]. Therefore, we predict that the new student would fail the course.
Bayesian vs. Frequentist
The Bayesian view of probability and the frequentist view of probability are the two major schools of thought in the field of statistics regarding how to interpret the probability of an event.
The Bayesian view of probability states that, for any event E, event E has a prior probability that represents how believable event E would occur prior to knowing anything about any other event whose occurrence could have an impact on event E's occurrence. Theoretically, this prior probability is a belief that represents the baseline probability for event E's occurrence. In practice, however, event E's prior probability is unknown, and therefore it must either be guessed at or be estimated using a sample of available data. After obtaining a guessed or estimated value of event E's prior probability, the Bayesian view holds that the probability, that is, the believability of event E's occurrence, can always be made more accurate should any new information regarding events that are relevant to event E become available. The Bayesian view also holds that the accuracy for the estimate of the probability of event E's occurrence is higher as long as there are more useful information available regarding events that are relevant to event E. The Bayesian view therefore holds that there is no intrinsic probability of occurrence associated with any event. If one adherers to the Bayesian view, one can then, for instance, predict tomorrow's weather as having a probability of, say, [math]\displaystyle{ \,50% }[/math] for rain. The Bayes classifier as described above is a good example of a classifier developed from the Bayesian view of probability. The earliest works that lay the framework for the Bayesian view of probability is accredited to Thomas Bayes (1702–1761).
In contrast to the Bayesian view of probability, the frequentist view of probability holds that there is an intrinsic probability of occurrence associated with every event to which one can carry out many, if not an infinite number, of well-defined independent random trials. In each trial for an event, the event either occurs or it does not occur. Suppose
[math]\displaystyle{ n_x }[/math] denotes the number of times that an event occurs during its trials and [math]\displaystyle{ n_t }[/math] denotes the total number of trials carried out for the event. The frequentist view of probability holds that, in the long run, where the number of trials for an event approaches infinity, one could theoretically approach the intrinsic value of the event's probability of occurrence to any arbitrary degree of accuracy, i.e., :[math]\displaystyle{ P(x) = \lim_{n_t\rightarrow \infty}\frac{n_x}{n_t} }[/math]. In practice, however, one can only carry out a finite number of trials for an event and, as a result, the probability of the event's occurrence can only be approximated as [math]\displaystyle{ P(x) \approx \frac{n_x}{n_t} }[/math]. If one adherers to the frequentist view, one cannot, for instance, predict the probability that there would be rain tomorrow. This is because one cannot possibly carry out trials for any event that is set in the future. The founder of the frequentist school of thought is arguably the famous Greek philosopher Aristotle. In his work Rhetoric, Aristotle gave the famous line "the probable is that which for the most part happens".
More information regarding the Bayesian and the frequentist schools of thought are available here. Furthermore, an interesting and informative youtube video that explains the Bayesian and frequentist views of probability is available here.
Linear and Quadratic Discriminant Analysis
First, we shall limit ourselves to the case where there are two classes, i.e. [math]\displaystyle{ \, \mathcal{Y}=\{0, 1\} }[/math]. In the above discussion, we introduced the Bayes classifier's decision boundary [math]\displaystyle{ \,D(h^*)=\{x: P(Y=1|X=x)=P(Y=0|X=x)\} }[/math], which represents a hyperplane that determines the class of any new data input depending on which side of the hyperplane the input lies in. Now, we shall look at how to derive the Bayes classifier's decision boundary under certain assumptions of the data. Linear discriminant analysis (LDA) and quadratic discriminant analysis (QDA) are two of the most well-known ways for deriving the Bayes classifier's decision boundary, and we shall look at each of them in turn.
Let us denote the likelihood [math]\displaystyle{ \ P(X=x|Y=y) }[/math] as [math]\displaystyle{ \ f_y(x) }[/math] and the prior probability [math]\displaystyle{ \ P(Y=y) }[/math] as [math]\displaystyle{ \ \pi_y }[/math].
First, we shall examine LDA. As explained above, the Bayes classifier is optimal. However, in practice, the prior and conditional densities are not known. Under LDA, one gets around this problem by making the assumptions that both of the two classes have multivariate normal (Gaussian) distributions and the two classes have the same covariance matrix [math]\displaystyle{ \, \Sigma }[/math]. Under the assumptions of LDA, we have: [math]\displaystyle{ \ P(X=x|Y=y) = f_y(x) = \frac{1}{ (2\pi)^{d/2}|\Sigma|^{1/2} }\exp\left( -\frac{1}{2} (x - \mu_k)^\top \Sigma^{-1} (x - \mu_k) \right) }[/math]. Now, to derive the Bayes classifier's decision boundary using LDA, we equate [math]\displaystyle{ \, P(Y=1|X=x) }[/math] to [math]\displaystyle{ \, P(Y=0|X=x) }[/math] and proceed from there. The derivation of [math]\displaystyle{ \,D(h^*) }[/math] is as follows:
- [math]\displaystyle{ \,Pr(Y=1|X=x)=Pr(Y=0|X=x) }[/math]
- [math]\displaystyle{ \,\Rightarrow \frac{Pr(X=x|Y=1)Pr(Y=1)}{Pr(X=x)}=\frac{Pr(X=x|Y=0)Pr(Y=0)}{Pr(X=x)} }[/math] (using Bayes' Theorem)
- [math]\displaystyle{ \,\Rightarrow Pr(X=x|Y=1)Pr(Y=1)=Pr(X=x|Y=0)Pr(Y=0) }[/math] (canceling the denominators)
- [math]\displaystyle{ \,\Rightarrow f_1(x)\pi_1=f_0(x)\pi_0 }[/math]
- [math]\displaystyle{ \,\Rightarrow \frac{1}{ (2\pi)^{d/2}|\Sigma|^{1/2} }\exp\left( -\frac{1}{2} (x - \mu_1)^\top \Sigma^{-1} (x - \mu_1) \right)\pi_1=\frac{1}{ (2\pi)^{d/2}|\Sigma|^{1/2} }\exp\left( -\frac{1}{2} (x - \mu_0)^\top \Sigma^{-1} (x - \mu_0) \right)\pi_0 }[/math]
- [math]\displaystyle{ \,\Rightarrow \exp\left( -\frac{1}{2} (x - \mu_1)^\top \Sigma^{-1} (x - \mu_1) \right)\pi_1=\exp\left( -\frac{1}{2} (x - \mu_0)^\top \Sigma^{-1} (x - \mu_0) \right)\pi_0 }[/math]
- [math]\displaystyle{ \,\Rightarrow -\frac{1}{2} (x - \mu_1)^\top \Sigma^{-1} (x - \mu_1) + \log(\pi_1)=-\frac{1}{2} (x - \mu_0)^\top \Sigma^{-1} (x - \mu_0) +\log(\pi_0) }[/math] (taking the log of both sides).
- [math]\displaystyle{ \,\Rightarrow \log(\frac{\pi_1}{\pi_0})-\frac{1}{2}\left( x^\top\Sigma^{-1}x + \mu_1^\top\Sigma^{-1}\mu_1 - 2x^\top\Sigma^{-1}\mu_1 - x^\top\Sigma^{-1}x - \mu_0^\top\Sigma^{-1}\mu_0 + 2x^\top\Sigma^{-1}\mu_0 \right)=0 }[/math] (expanding out)
- [math]\displaystyle{ \,\Rightarrow \log(\frac{\pi_1}{\pi_0})-\frac{1}{2}\left( \mu_1^\top\Sigma^{-1} \mu_1-\mu_0^\top\Sigma^{-1}\mu_0 - 2x^\top\Sigma^{-1}(\mu_1-\mu_0) \right)=0 }[/math] (canceling out alike terms and factoring).
It is easy to see that, under LDA, the Bayes's classifier's decision boundary [math]\displaystyle{ \,D(h^*) }[/math] has the form [math]\displaystyle{ \,ax+b=0 }[/math] and it is linear in [math]\displaystyle{ \,x }[/math]. This is where the word linear in linear discriminant analysis comes from.
LDA under the two-classes case can easily be generalized to the general case where there are [math]\displaystyle{ \,k \ge 2 }[/math] classes. In the general case, suppose we wish to find the Bayes classifier's decision boundary between the two classes [math]\displaystyle{ \,m }[/math] and [math]\displaystyle{ \,n }[/math], then all we need to do is follow a derivation very similar to the one shown above, except with the classes [math]\displaystyle{ \,1 }[/math] and [math]\displaystyle{ \,0 }[/math] being replaced by the classes [math]\displaystyle{ \,m }[/math] and [math]\displaystyle{ \,n }[/math]. Following through with a similar derivation as the one shown above, one obtains the Bayes classifier's decision boundary [math]\displaystyle{ \,D(h^*) }[/math] between classes [math]\displaystyle{ \,m }[/math] and [math]\displaystyle{ \,n }[/math] to be [math]\displaystyle{ \,\log(\frac{\pi_m}{\pi_n})-\frac{1}{2}\left( \mu_m^\top\Sigma^{-1}
\mu_m-\mu_n^\top\Sigma^{-1}\mu_n - 2x^\top\Sigma^{-1}(\mu_m-\mu_n) \right)=0 }[/math] . In addition, for any two classes [math]\displaystyle{ \,m }[/math] and [math]\displaystyle{ \,n }[/math] for whom we would like to find the Bayes classifier's decision boundary using LDA, if [math]\displaystyle{ \,m }[/math] and [math]\displaystyle{ \,n }[/math] both have the same number of data, then, in this special case, the resulting decision boundary would lie exactly halfway between the centers (means) of [math]\displaystyle{ \,m }[/math] and [math]\displaystyle{ \,n }[/math].
The Bayes classifier's decision boundary for any two classes as derived using LDA looks something like the one that can be found in this link:
Although the assumption under LDA may not hold true for most real-world data, it nevertheless usually performs quite well in practice, where it often provides near-optimal classifications. For instance, the Z-Score credit risk model that was designed by Edward Altman in 1968 and revisited in 2000, is essentially a weighted LDA. This model has demonstrated a 85-90% success rate in predicting bankruptcy, and for this reason it is still in use today.
According to this link, some of the limitations of LDA include:
- LDA implicitly assumes that the data in each class has a Gaussian distribution.
- LDA implicitly assumes that the mean rather than the variance is the discriminating factor.
- LDA may over-fit the training data.
Linear and Quadratic Discriminant Analysis cont'd - September 23, 2010
LDA x QDA
Linear discriminant analysis[1] is a statistical method used to find the linear combination of features which best separate two or more classes of objects or events. It is widely applied in classifying diseases, positioning, product management, and marketing research. LDA assumes that the different classes have the same covariance matrix [math]\displaystyle{ \, \Sigma }[/math].
Quadratic Discriminant Analysis[2], on the other hand, aims to find the quadratic combination of features. It is more general than linear discriminant analysis. Unlike LDA, QDA does not make the assumption that the different classes have the same covariance matrix [math]\displaystyle{ \, \Sigma }[/math]. Instead, QDA makes the assumption that each class [math]\displaystyle{ \, k }[/math] has its own covariance matrix [math]\displaystyle{ \, \Sigma_k }[/math].
The derivation of the Bayes classifier's decision boundary [math]\displaystyle{ \,D(h^*) }[/math] under QDA is similar to that under LDA. Again, let us first consider the two-classes case where [math]\displaystyle{ \, \mathcal{Y}=\{0, 1\} }[/math]. This derivation is given as follows:
- [math]\displaystyle{ \,Pr(Y=1|X=x)=Pr(Y=0|X=x) }[/math]
- [math]\displaystyle{ \,\Rightarrow \frac{Pr(X=x|Y=1)Pr(Y=1)}{Pr(X=x)}=\frac{Pr(X=x|Y=0)Pr(Y=0)}{Pr(X=x)} }[/math] (using Bayes' Theorem)
- [math]\displaystyle{ \,\Rightarrow Pr(X=x|Y=1)Pr(Y=1)=Pr(X=x|Y=0)Pr(Y=0) }[/math] (canceling the denominators)
- [math]\displaystyle{ \,\Rightarrow f_1(x)\pi_1=f_0(x)\pi_0 }[/math]
- [math]\displaystyle{ \,\Rightarrow \frac{1}{ (2\pi)^{d/2}|\Sigma_1|^{1/2} }\exp\left( -\frac{1}{2} (x - \mu_1)^\top \Sigma_1^{-1} (x - \mu_1) \right)\pi_1=\frac{1}{ (2\pi)^{d/2}|\Sigma_0|^{1/2} }\exp\left( -\frac{1}{2} (x - \mu_0)^\top \Sigma_0^{-1} (x - \mu_0) \right)\pi_0 }[/math]
- [math]\displaystyle{ \,\Rightarrow \frac{1}{|\Sigma_1|^{1/2} }\exp\left( -\frac{1}{2} (x - \mu_1)^\top \Sigma_1^{-1} (x - \mu_1) \right)\pi_1=\frac{1}{|\Sigma_0|^{1/2} }\exp\left( -\frac{1}{2} (x - \mu_0)^\top \Sigma_0^{-1} (x - \mu_0) \right)\pi_0 }[/math] (by cancellation)
- [math]\displaystyle{ \,\Rightarrow -\frac{1}{2}\log(|\Sigma_1|)-\frac{1}{2} (x - \mu_1)^\top \Sigma_1^{-1} (x - \mu_1)+\log(\pi_1)=-\frac{1}{2}\log(|\Sigma_0|)-\frac{1}{2} (x - \mu_0)^\top \Sigma_0^{-1} (x - \mu_0)+\log(\pi_0) }[/math] (by taking the log of both sides)
- [math]\displaystyle{ \,\Rightarrow \log(\frac{\pi_1}{\pi_0})-\frac{1}{2}\log(\frac{|\Sigma_1|}{|\Sigma_0|})-\frac{1}{2}\left( x^\top\Sigma_1^{-1}x + \mu_1^\top\Sigma_1^{-1}\mu_1 - 2x^\top\Sigma_1^{-1}\mu_1 - x^\top\Sigma_0^{-1}x - \mu_0^\top\Sigma_0^{-1}\mu_0 + 2x^\top\Sigma_0^{-1}\mu_0 \right)=0 }[/math] (by expanding out)
- [math]\displaystyle{ \,\Rightarrow \log(\frac{\pi_1}{\pi_0})-\frac{1}{2}\log(\frac{|\Sigma_1|}{|\Sigma_0|})-\frac{1}{2}\left( x^\top(\Sigma_1^{-1}-\Sigma_0^{-1})x + \mu_1^\top\Sigma_1^{-1}\mu_1 - \mu_0^\top\Sigma_0^{-1}\mu_0 - 2x^\top(\Sigma_1^{-1}\mu_1-\Sigma_0^{-1}\mu_0) \right)=0 }[/math]
It is easy to see that, under QDA, the decision boundary [math]\displaystyle{ \,D(h^*) }[/math] has the form [math]\displaystyle{ \,ax^2+bx+c=0 }[/math] and it is quadratic in [math]\displaystyle{ \,x }[/math]. This is where the word quadratic in quadratic discriminant analysis comes from.
As is the case with LDA, QDA under the two-classes case can easily be generalized to the general case where there are [math]\displaystyle{ \,k \ge 2 }[/math] classes. In the general case, suppose we wish to find the Bayes classifier's decision boundary between the two classes [math]\displaystyle{ \,m }[/math] and [math]\displaystyle{ \,n }[/math], then all we need to do is follow a derivation very similar to the one shown above, except with the classes [math]\displaystyle{ \,1 }[/math] and [math]\displaystyle{ \,0 }[/math] being replaced by the classes [math]\displaystyle{ \,m }[/math] and [math]\displaystyle{ \,n }[/math]. Following through with a similar derivation as the one shown above, one obtains the Bayes classifier's decision boundary [math]\displaystyle{ \,D(h^*) }[/math] between classes [math]\displaystyle{ \,m }[/math] and [math]\displaystyle{ \,n }[/math] to be [math]\displaystyle{ \,\log(\frac{\pi_m}{\pi_n})-\frac{1}{2}\log(\frac{|\Sigma_m|}{|\Sigma_n|})-\frac{1}{2}\left( x^\top(\Sigma_m^{-1}-\Sigma_n^{-1})x + \mu_m^\top\Sigma_m^{-1}\mu_m - \mu_n^\top\Sigma_n^{-1}\mu_n - 2x^\top(\Sigma_m^{-1}\mu_m-\Sigma_n^{-1}\mu_n) \right)=0 }[/math].
Summarizing LDA and QDA
We can summarize what we have learned so far into the following theorem.
Theorem:
Suppose that [math]\displaystyle{ \,Y \in \{1,\dots,K\} }[/math], if [math]\displaystyle{ \,f_k(x) = Pr(X=x|Y=k) }[/math] is Gaussian, the Bayes Classifier rule is
- [math]\displaystyle{ \,h^*(x) = \arg\max_{k} \delta_k(x) }[/math]
where,
- In the case of LDA, which assumes that a common covariance matrix is shared by all classes, [math]\displaystyle{ \,\delta_k(x) = x^\top\Sigma^{-1}\mu_k - \frac{1}{2}\mu_k^\top\Sigma^{-1}\mu_k + log (\pi_k) }[/math], and the Bayes classifier's decision boundary [math]\displaystyle{ \,D(h^*) }[/math] is linear in [math]\displaystyle{ \,x }[/math].
- In the case of QDA, which assumes that each class has its own covariance matrix, [math]\displaystyle{ \,\delta_k(x) = - \frac{1}{2}log(|\Sigma_k|) - \frac{1}{2}(x-\mu_k)^\top\Sigma_k^{-1}(x-\mu_k) + log (\pi_k) }[/math], and the Bayes classifier's decision boundary [math]\displaystyle{ \,D(h^*) }[/math] is quadratic in [math]\displaystyle{ \,x }[/math].
Note [math]\displaystyle{ \,\arg\max_{k} \delta_k(x) }[/math]returns the set of k for which [math]\displaystyle{ \,\delta_k(x) }[/math] attains its largest value.
In practice
We need to estimate the prior, so in order to do this, we use the sample estimates for [math]\displaystyle{ \,\pi,\mu_k,\Sigma_k }[/math] in place of their true values, i.e.
[math]\displaystyle{ \,\hat{\pi_k} = \hat{Pr}(y=k) = \frac{n_k}{n} }[/math]
[math]\displaystyle{ \,\hat{\mu_k} = \frac{1}{n_k}\sum_{i:y_i=k}x_i }[/math]
[math]\displaystyle{ \,\hat{\Sigma_k} = \frac{1}{n_k-d}\sum_{i:y_i=k}(x_i-\hat{\mu_k})(x_i-\hat{\mu_k})^\top }[/math]
Common covariance, denoted [math]\displaystyle{ \Sigma }[/math], is defined as the weighted average of the covariance for each class.
In the case where we need a common covariance matrix, we get the estimate using the following equation:
[math]\displaystyle{ \,\Sigma=\frac{\sum_{r=1}^{k}(n_r\Sigma_r)}{n} }[/math]
Where: [math]\displaystyle{ \,n_r }[/math] is the number of data points in class r, [math]\displaystyle{ \,\Sigma_r }[/math] is the covariance of class r and [math]\displaystyle{ \,n }[/math] is the total number of data points.
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: Is the common covariance really a maximum likelihood estimate? Does it really matter if it is? I know the above 3 "sample estimates" are in fact maximum likelihood estimates, perhaps the following statement should be moved up to describe them. Please improve this article if you can. (October 14 2010) | small = | smallimage = | smallimageright = | smalltext = }}
This is a Maximum Likelihood estimate.
Computation For QDA And LDA
First, let us consider QDA, and examine each of the following two cases regarding QDA.
Case 1: (Example) [math]\displaystyle{ \, \Sigma_k = I }[/math]
[math]\displaystyle{ \, \Sigma_k = I }[/math] for every class [math]\displaystyle{ \,k }[/math] implies that our data is spherical. This means that the data of each class [math]\displaystyle{ \,k }[/math] is distributed symmetrically around the center [math]\displaystyle{ \,\mu_k }[/math], i.e. the isocontours are all circles.
We have:
[math]\displaystyle{ \,\delta_k = - \frac{1}{2}log(|I|) - \frac{1}{2}(x-\mu_k)^\top I(x-\mu_k) + log (\pi_k) }[/math]
We see that the first term in the above equation, [math]\displaystyle{ \,\frac{-1}{2}log(|I|) }[/math], is zero since [math]\displaystyle{ \ |I|=1 }[/math]. The second term contains [math]\displaystyle{ \, (x-\mu_k)^\top I(x-\mu_k) = (x-\mu_k)^\top(x-\mu_k) }[/math], which is the squared Euclidean distance between [math]\displaystyle{ \,x }[/math] and [math]\displaystyle{ \,\mu_k }[/math]. Therefore we can find the distance between a point and each center and adjust it with the log of the prior, [math]\displaystyle{ \,log(\pi_k) }[/math]. The class that has the minimum distance will maximize [math]\displaystyle{ \,\delta_k }[/math]. According to the theorem, we can then classify the point to a specific class [math]\displaystyle{ \,k }[/math].
Case 2: (General Case) [math]\displaystyle{ \, \Sigma_k \ne I }[/math]
We can decompose this as:
[math]\displaystyle{ \, \Sigma_k = U_kS_kV_k^\top = U_kS_kU_k^\top }[/math] (In general when [math]\displaystyle{ \,X=U_kS_kV_k^\top }[/math], [math]\displaystyle{ \,U_k }[/math] is the eigenvectors of [math]\displaystyle{ \,X_kX_k^T }[/math] and [math]\displaystyle{ \,V_k }[/math] is the eigenvectors of [math]\displaystyle{ \,X_k^\top X_k }[/math]. So if [math]\displaystyle{ \, X_k }[/math] is symmetric, we will have [math]\displaystyle{ \, U_k=V_k }[/math]. Here [math]\displaystyle{ \, \Sigma_k }[/math] is symmetric, because it is the covariance matrix of [math]\displaystyle{ X_k }[/math]) and the inverse of [math]\displaystyle{ \,\Sigma_k }[/math] is
[math]\displaystyle{ \, \Sigma_k^{-1} = (U_kS_kU_k^\top)^{-1} = (U_k^\top)^{-1}S_k^{-1}U_k^{-1} = U_kS_k^{-1}U_k^\top }[/math] (since [math]\displaystyle{ \,U_k }[/math] is orthonormal)
So from the formula for [math]\displaystyle{ \,\delta_k }[/math], the second term is
- [math]\displaystyle{ \begin{align} (x-\mu_k)^\top\Sigma_k^{-1}(x-\mu_k)&= (x-\mu_k)^\top U_kS_k^{-1}U_k^T(x-\mu_k)\\ & = (U_k^\top x-U_k^\top\mu_k)^\top S_k^{-1}(U_k^\top x-U_k^\top \mu_k)\\ & = (U_k^\top x-U_k^\top\mu_k)^\top S_k^{-\frac{1}{2}}S_k^{-\frac{1}{2}}(U_k^\top x-U_k^\top\mu_k) \\ & = (S_k^{-\frac{1}{2}}U_k^\top x-S_k^{-\frac{1}{2}}U_k^\top\mu_k)^\top I(S_k^{-\frac{1}{2}}U_k^\top x-S_k^{-\frac{1}{2}}U_k^\top \mu_k) \\ & = (S_k^{-\frac{1}{2}}U_k^\top x-S_k^{-\frac{1}{2}}U_k^\top\mu_k)^\top(S_k^{-\frac{1}{2}}U_k^\top x-S_k^{-\frac{1}{2}}U_k^\top \mu_k) \\ \end{align} }[/math]
where we have the squared Euclidean distance between [math]\displaystyle{ \, S_k^{-\frac{1}{2}}U_k^\top x }[/math] and [math]\displaystyle{ \, S_k^{-\frac{1}{2}}U_k^\top\mu_k }[/math].
A transformation of all the data points can be done from [math]\displaystyle{ \,x }[/math] to [math]\displaystyle{ \,x^* }[/math] where [math]\displaystyle{ \, x^* \leftarrow S_k^{-\frac{1}{2}}U_k^\top x }[/math].
A similar transformation of all the centers can be done from [math]\displaystyle{ \,\mu_k }[/math] to [math]\displaystyle{ \,\mu_k^* }[/math] where [math]\displaystyle{ \, \mu_k^* \leftarrow S_k^{-\frac{1}{2}}U_k^\top \mu_k }[/math].
It is now possible to do classification with [math]\displaystyle{ \,x^* }[/math] and [math]\displaystyle{ \,\mu_k^* }[/math], treating them as in Case 1 above.
Note that when we have multiple classes, they must all have the same transformation, else, ahead of time we would have to assume a data point belongs to one class or the other. All classes therefore need to have the same shape for classification to be applicable using this method. So this method works for LDA.
If the classes have different shapes, in another word, have different covariance [math]\displaystyle{ \,\Sigma_k }[/math], can we use the same method to transform all data points [math]\displaystyle{ \,x }[/math] to [math]\displaystyle{ \,x^* }[/math]?
The answer is NO. Consider that you have two classes with different shapes, then consider transforming them to the same shape. Given a data point, justify which class this point belongs to. The question is, which transformation can you use? For example, if you use the transformation of class A, then you have assumed that this data point belongs to class A.
In summary, to apply QDA on a data set [math]\displaystyle{ \,X }[/math], in the general case where [math]\displaystyle{ \, \Sigma_k \ne I }[/math] for each class [math]\displaystyle{ \,k }[/math], one can proceed as follows:
- Step 1: For each class [math]\displaystyle{ \,k }[/math], apply singular value decomposition on [math]\displaystyle{ \,X_k }[/math] to obtain [math]\displaystyle{ \,S_k }[/math] and [math]\displaystyle{ \,U_k }[/math].
- Step 2: For each class [math]\displaystyle{ \,k }[/math], transform each [math]\displaystyle{ \,x }[/math] belonging to that class to [math]\displaystyle{ \,x^* = S_k^{-\frac{1}{2}}U_k^\top x }[/math], and transform its center [math]\displaystyle{ \,\mu_k }[/math] to [math]\displaystyle{ \,\mu_k^* = S_k^{-\frac{1}{2}}U_k^\top \mu_k }[/math].
- Step 3: For each data point [math]\displaystyle{ \,x \in X }[/math], find the squared Euclidean distance between the transformed data point [math]\displaystyle{ \,x^* }[/math] and the transformed center [math]\displaystyle{ \,\mu^* }[/math] of each class, and assign [math]\displaystyle{ \,x }[/math] to the class such that the squared Euclidean distance between [math]\displaystyle{ \,x^* }[/math] and [math]\displaystyle{ \,\mu^* }[/math] is the least over all of the classes.
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: If someone can provide a derivation for LDA, that would be great. Thanks. Please improve this article if you can. (October 8 2010) | small = | smallimage = | smallimageright = | smalltext = }}
Now, let us consider LDA.
In the case of LDA, one can derive a classification scheme that is quite similar to that derived from Case 2 of QDA shown above. To apply LDA on a data set [math]\displaystyle{ \,X }[/math], one can proceed as follows:
- Step 1: Apply singular value decomposition on [math]\displaystyle{ \,X }[/math] to obtain [math]\displaystyle{ \,S }[/math] and [math]\displaystyle{ \,U }[/math].
- Step 2: For each [math]\displaystyle{ \,x \in X }[/math], transform [math]\displaystyle{ \,x }[/math] to [math]\displaystyle{ \,x^* = S^{-\frac{1}{2}}U^\top x }[/math], and transform each center [math]\displaystyle{ \,\mu }[/math] to [math]\displaystyle{ \,\mu^* = S^{-\frac{1}{2}}U^\top \mu }[/math].
- Step 3: For each data point [math]\displaystyle{ \,x \in X }[/math], find the squared Euclidean distance between the transformed data point [math]\displaystyle{ \,x^* }[/math] and the transformed center [math]\displaystyle{ \,\mu^* }[/math] of each class, and assign [math]\displaystyle{ \,x }[/math] to the class such that the squared Euclidean distance between [math]\displaystyle{ \,x^* }[/math] and [math]\displaystyle{ \,\mu^* }[/math] is the least over all of the classes.
Kernel QDA
In actual data scenarios, it is generally true that QDA will provide a better classifier for the data then LDA because QDA does not assume that the covariance matrix for each class is identical, as LDA assumes. However, QDA still assumes that the class conditional distribution is Gaussian, which is not always the case in real-life scenarios. The link provided at the beginning of this paragraph describes a kernel-based QDA method which does not have the Gaussian distribution assumption.
The Number of Parameters in LDA and QDA
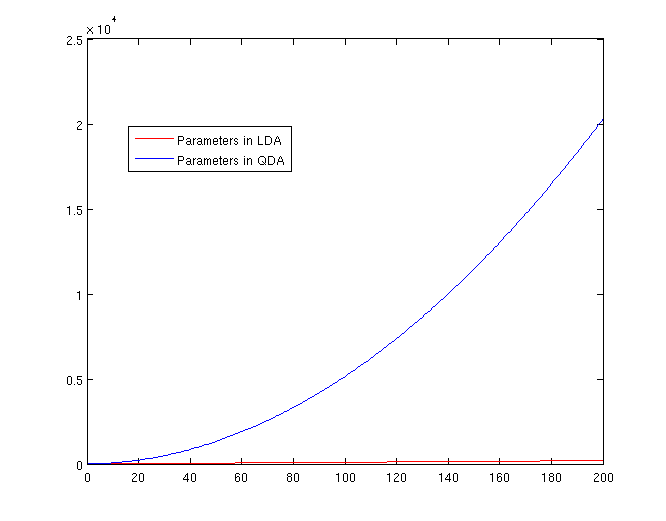
Both LDA and QDA require us to estimate parameters. The more estimation we have to do, the less robust our classification algorithm will be.
LDA: Since we just need to compare the differences between one given class and remaining [math]\displaystyle{ \,K-1 }[/math] classes, totally, there are [math]\displaystyle{ \,K-1 }[/math] differences. For each of them, [math]\displaystyle{ \,a^{T}x+b }[/math] requires [math]\displaystyle{ \,d+1 }[/math] parameters. Therefore, there are [math]\displaystyle{ \,(K-1)\times(d+1) }[/math] parameters.
QDA: For each of the differences, [math]\displaystyle{ \,x^{T}ax + b^{T}x + c }[/math] requires [math]\displaystyle{ \frac{1}{2}(d+1)\times d + d + 1 = \frac{d(d+3)}{2}+1 }[/math] parameters. Therefore, there are [math]\displaystyle{ (K-1)(\frac{d(d+3)}{2}+1) }[/math] parameters.
Trick: Using LDA to do QDA - September 28, 2010
There is a trick that allows us to use the linear discriminant analysis (LDA) algorithm to generate as its output a quadratic function that can be used to classify data. This trick is similar to, but more primitive than, the Kernel trick that will be discussed later in the course.
Essentially, the trick involves adding one or more new features (i.e. new dimensions) that just contain our original data projected to that dimension. We then do LDA on our new higher-dimensional data. The answer provided by LDA can then be collapsed onto a lower dimension, giving us a quadratic answer.
Motivation
Why would we want to use LDA over QDA? In situations where we have fewer data points, LDA turns out to be more robust.
If we look back at the equations for LDA and QDA, we see that in LDA we must estimate [math]\displaystyle{ \,\mu_1 }[/math], [math]\displaystyle{ \,\mu_2 }[/math] and [math]\displaystyle{ \,\Sigma }[/math]. In QDA we must estimate all of those, plus another [math]\displaystyle{ \,\Sigma }[/math]; the extra [math]\displaystyle{ \,\frac{d(d-1)}{2} }[/math] estimations make QDA less robust with fewer data points.
Theoretically
Suppose we can estimate some vector [math]\displaystyle{ \underline{w}^T }[/math] such that
[math]\displaystyle{ y = \underline{w}^Tx }[/math]
where [math]\displaystyle{ \underline{w} }[/math] is a d-dimensional column vector, and [math]\displaystyle{ x\ \epsilon\ \mathbb{R}^d }[/math] (vector in d dimensions).
We also have a non-linear function [math]\displaystyle{ g(x) = y = x^Tvx + \underline{w}^Tx }[/math] that we cannot estimate.
Using our trick, we create two new vectors, [math]\displaystyle{ \,\underline{w}^* }[/math] and [math]\displaystyle{ \,x^* }[/math] such that:
[math]\displaystyle{ \underline{w}^{*T} = [w_1,w_2,...,w_d,v_1,v_2,...,v_d] }[/math]
and
[math]\displaystyle{ x^{*T} = [x_1,x_2,...,x_d,{x_1}^2,{x_2}^2,...,{x_d}^2] }[/math]
We can then estimate a new function, [math]\displaystyle{ g^*(x,x^2) = y^* = \underline{w}^{*T}x^* }[/math].
Note that we can do this for any [math]\displaystyle{ x }[/math] and in any dimension; we could extend a [math]\displaystyle{ D \times n }[/math] matrix to a quadratic dimension by appending another [math]\displaystyle{ D \times n }[/math] matrix with the original matrix squared, to a cubic dimension with the original matrix cubed, or even with a different function altogether, such as a [math]\displaystyle{ \,sin(x) }[/math] dimension. Pay attention, We don't do QDA with LDA. If we try QDA directly on this problem the resulting decision boundary will be different. Here we try to find a nonlinear boundary for a better possible boundary but it is different with general QDA method. We can call it nonlinear LDA.
By Example
Let's use our trick to do a quadratic analysis of the 2_3 data using LDA.
>> load 2_3; >> [U, sample] = princomp(X'); >> sample = sample(:,1:2);
- We start off the same way, by using PCA to reduce the dimensionality of our data to 2.
>> X_star = zeros(400,4);
>> X_star(:,1:2) = sample(:,:);
>> for i=1:400
for j=1:2
X_star(i,j+2) = X_star(i,j)^2;
end
end
- This projects our sample into two more dimensions by squaring our initial two dimensional data set.
>> group = ones(400,1); >> group(201:400) = 2; >> [class, error, POSTERIOR, logp, coeff] = classify(X_star, X_star, group, 'linear'); >> sum (class==group) ans = 375
- We can now display our results.
>> k = coeff(1,2).const;
>> l = coeff(1,2).linear;
>> f = sprintf('0 = %g+%g*x+%g*y+%g*(x)^2+%g*(y)^2', k, l(1), l(2),l(3),l(4));
>> ezplot(f,[min(sample(:,1)), max(sample(:,1)), min(sample(:,2)), max(sample(:,2))]);
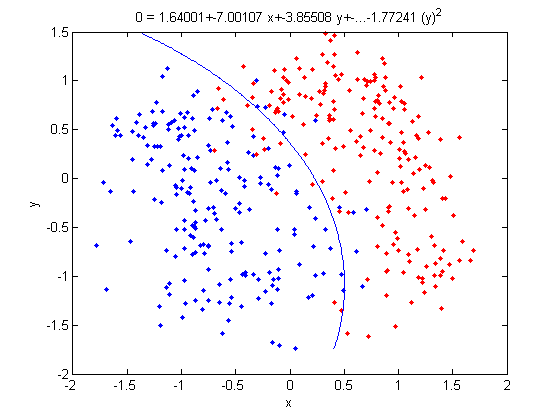
- Not only does LDA give us a better result than it did previously, it actually beats QDA, which only correctly classified 371 data points for this data set. Continuing this procedure by adding another two dimensions with [math]\displaystyle{ x^4 }[/math] (i.e. we set
X_star(i,j+2) = X_star(i,j)^4) we can correctly classify 376 points.
LDA and QDA in Matlab
We have examined the theory behind Linear Discriminant Analysis (LDA) and Quadratic Discriminant Analysis (QDA) above; how do we use these algorithms in practice? Matlab offers us a function called classify that allows us to perform LDA and QDA quickly and easily.
In class, we were shown an example of using LDA and QDA on the 2_3 data that is used in the first assignment. The code below applies LDA to the same data set and reproduces that example, slightly modified, and explains each step.
>> load 2_3; >> [U, sample] = princomp(X'); >> sample = sample(:,1:2);
- First, we do principal component analysis (PCA) on the 2_3 data to reduce the dimensionality of the original data from 64 dimensions to 2. Doing this makes it much easier to visualize the results of the LDA and QDA algorithms.
>> plot (sample(1:200,1), sample(1:200,2), '.'); >> hold on; >> plot (sample(201:400,1), sample(201:400,2), 'r.');
- Recall that in the 2_3 data, the first 200 elements are images of the number two handwritten and the last 200 elements are images of the number three handwritten. This code sets up a plot of the data such that the points that represent a 2 are blue, while the points that represent a 3 are red.
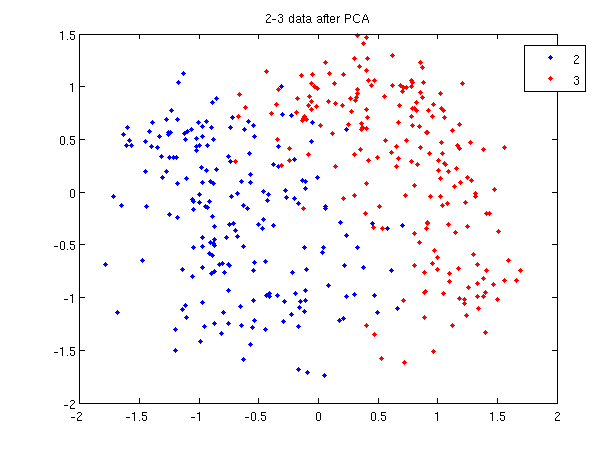
title and legend for information on adding the title and legend.- Before using
classifywe can set up a vector that contains the actual labels for our data, to train the classification algorithm. If we don't know the labels for the data, then the element in thegroupvector should be an empty string orNaN. (See grouping data for more information.)
>> group = ones(400,1); >> group(201:400) = 2;
- We can now classify our data.
>> [class, error, POSTERIOR, logp, coeff] = classify(sample, sample, group, 'linear');
- The full details of this line can be examined in the Matlab help file linked above. What we care about are
class, which contains the labels that the algorithm thinks that each data point belongs to, andcoeff, which contains information about the line that the algorithm created to separate the data into the two classes.
- We can see the efficacy of the algorithm by comparing
classtogroup.
>> sum (class==group) ans = 369
- This compares the value in
classto the value ingroup. The answer of 369 tells us that the algorithm correctly determined the classes of the points 369 times, out of a possible 400 data points. This gives us an empirical error rate of 0.0775.
- We can see the line produced by LDA using
coeff.
>> k = coeff(1,2).const;
>> l = coeff(1,2).linear;
>> f = sprintf('0 = %g+%g*x+%g*y', k, l(1), l(2));
>> ezplot(f, [min(sample(:,1)), max(sample(:,1)), min(sample(:,2)), max(sample(:,2))]);
- Those familiar with the programming language C will find the
sprintfline refreshingly familiar; those with no exposure to C are directed to Matlab'ssprintfpage. Essentially, this code sets up the equation of the line in the form0 = a + bx + cy. We then use theezplotfunction to plot the line.
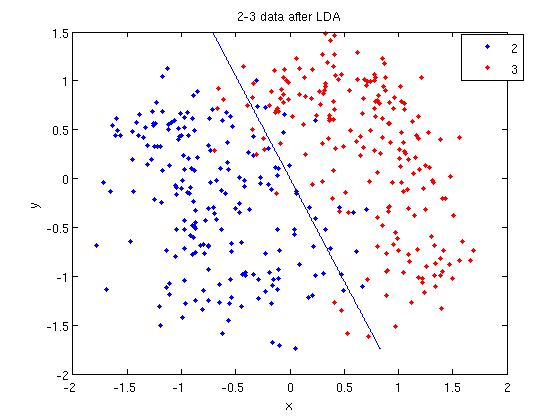
- Let's perform the same steps, except this time using QDA. The main difference with QDA is a slightly different call to
classify, and a more complicated procedure to plot the line.
>> [class, error, POSTERIOR, logp, coeff] = classify(sample, sample, group, 'quadratic');
>> sum (class==group)
ans =
371
>> k = coeff(1,2).const;
>> l = coeff(1,2).linear;
>> q = coeff(1,2).quadratic;
>> f = sprintf('0 = %g+%g*x+%g*y+%g*x^2+%g*x*y+%g*y^2', k, l(1), l(2), q(1,1), q(1,2)+q(2,1), q(2,2));
>> ezplot(f, [min(sample(:,1)), max(sample(:,1)), min(sample(:,2)), max(sample(:,2))]);
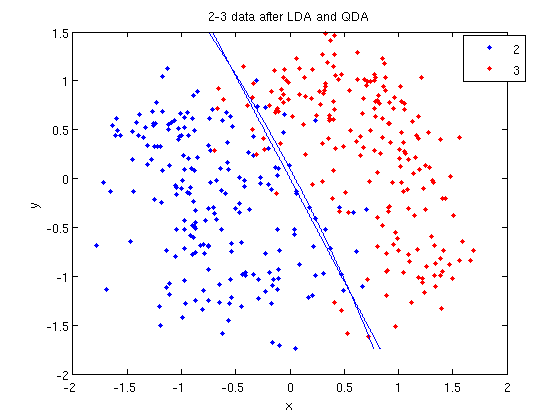
classify can also be used with other discriminant analysis algorithms. The steps laid out above would only need to be modified slightly for those algorithms.
Recall: An analysis of the function of princomp in matlab.
In our assignment 1, we have learnt that how to perform Principal Component Analysis using SVD method. In fact, the matlab offers us a function called princomp which can perform PCA conveniently. From the matlab help file on princomp, you can find the details about this function. But here we will analyze the code of the function of princomp() in matlab to find something different when comparing with SVD method. The following is the code of princomp and explanations to some emphasized steps.
function [pc, score, latent, tsquare] = princomp(x);
% PRINCOMP Principal Component Analysis (centered and scaled data).
% [PC, SCORE, LATENT, TSQUARE] = PRINCOMP(X) takes a data matrix X and
% returns the principal components in PC, the so-called Z-scores in SC
% ORES, the eigenvalues of the covariance matrix of X in LATENT,
% and Hotelling's T-squared statistic for each data point in TSQUARE.
% Reference: J. Edward Jackson, A User's Guide to Principal Components
% John Wiley & Sons, Inc. 1991 pp. 1-25.
% B. Jones 3-17-94
% Copyright 1993-2002 The MathWorks, Inc.
% $Revision: 2.9 $ $Date: 2002/01/17 21:31:45 $
[m,n] = size(x); % get the lengh of the rows and columns of matrix x.
r = min(m-1,n); % max possible rank of X
avg = mean(x); % the mean of every column of X
centerx = (x - avg(ones(m,1),:));
% centers X by subtracting off column means
[U,latent,pc] = svd(centerx./sqrt(m-1),0);
% "economy size" decomposition
score = centerx*pc;
% the representation of X in the principal component space
if nargout < 3
return;
end
latent = diag(latent).^2;
if (r latent = [latent(1:r); zeros(n-r,1)];
score(:,r+1:end) = 0;
end
if nargout < 4
return;
end
tmp = sqrt(diag(1./latent(1:r)))*score(:,1:r)';
tsquare = sum(tmp.*tmp)';
From the above code, we should pay attention to the following aspects when comparing with SVD method:
First, Rows of [math]\displaystyle{ \,X }[/math] correspond to observations, columns to variables. When using princomp on 2_3 data in assignment 1, note that we take the transpose of [math]\displaystyle{ \,X }[/math].
>> load 2_3; >> [U, score] = princomp(X');
Second, princomp centers X by subtracting off column means.
The third, when [math]\displaystyle{ \,X=UdV' }[/math], princomp uses [math]\displaystyle{ \,V }[/math] as coefficients for principal components, rather than [math]\displaystyle{ \,U }[/math].
The following is an example to perform PCA using princomp and SVD respectively to get the same results.
- SVD method
>> load 2_3 >> mn=mean(X,2); >> X1=X-repmat(mn,1,400); >> [s d v]=svd(X1'); >> y=X1'*v;
- princomp
>>[U score]=princomp(X');
Then we can see that y=score, v=U.
useful resouces: LDA and QDA in Matlab[3],[4],[5]
Reference
1. Harry Zhang. The optimality of naive bayes. FLAIRS Conference. AAAI Press, 2004
2. Rich Caruana and Alexandru N. Mizil. An empirical comparison of supervised learning algorithms. In ICML ’06: Proceedings of the 23rd international conference on Machine learning, pages 161–168, New York, NY, USA, 2006, ACM.
Related links to LDA & QDA
LDA:[6]
Regularized linear discriminant analysis and its application in microarrays
MATHEMATICAL OPERATIONS OF LDA
Application in face recognition and in market
QDA:[8]
Principal Component Analysis - September 30, 2010
Rough definition
Keepings two important aspects of data analysis in mind:
- Reducing covariance in data
- Preserving information stored in data(Variance is a source of information)
Principal component analysis (PCA) is a dimensionality-reduction method invented by Karl Pearson in 1901 [9]. Depending on where this methodology is applied, other common names of PCA include the Karhunen–Loève transform (KLT) , the Hotelling transform, and the proper orthogonal decomposition (POD). PCA is the simplist eigenvector-based multivariate analysis. It reduces the dimensionality of the data by revealing the internal structure of the data in a way that best explains the variance in the data. To this end, PCA works by using a user-defined number of the most important directions of variation (dimensions or principal components) of the data to project the data onto these directions so as to produce a lower-dimensional representation of the original data. The resulting lower-dimensional representation of our data is usually much easier to visualize and it also exhibits the most informative aspects (dimensions) of our data whilst capturing as much of the variation exhibited by our data as it possibly could.
Furthermore, if one considers the lower dimensional representation produced by PCA as a least squares fit of our original data, then it can also be easily shown that this representation is the one that minimizes the reconstruction error of our data. It should be noted however, that one usually does not have control over which dimensions PCA deems to be the most informative for a given set of data, and thus one usually does not know which dimensions PCA selects to be the most informative dimensions in order to create the lower-dimensional representation.
Suppose [math]\displaystyle{ \,X }[/math] is our data matrix containing [math]\displaystyle{ \,d }[/math]-dimensional data. The idea behind PCA is to apply singular value decomposition to [math]\displaystyle{ \,X }[/math] to replace the rows of [math]\displaystyle{ \,X }[/math] by a subset of it that captures as much of the variance in [math]\displaystyle{ \,X }[/math] as possible. First, through the application of singular value decomposition to [math]\displaystyle{ \,X }[/math], PCA obtains all of our data's directions of variation. These directions would also be ordered from left to right, with the leftmost directions capturing the most amount of variation in our data and the rightmost directions capturing the least amount. Then, PCA uses a subset of these directions to map our data from its original space to a lower-dimensional space.
By applying singular value decomposition to [math]\displaystyle{ \,X }[/math], [math]\displaystyle{ \,X }[/math] is decomposed as [math]\displaystyle{ \,X = U\Sigma V^T \, }[/math]. The [math]\displaystyle{ \,d }[/math] columns of [math]\displaystyle{ \,U }[/math] are the eigenvectors of [math]\displaystyle{ \,XX^T \, }[/math].
The [math]\displaystyle{ \,d }[/math] columns of [math]\displaystyle{ \,V }[/math] are the eigenvectors of [math]\displaystyle{ \,X^TX \, }[/math]. The [math]\displaystyle{ \,d }[/math] diagonal values of [math]\displaystyle{ \,\Sigma }[/math] are the square roots of the eigenvalues of [math]\displaystyle{ \,XX^T \, }[/math] (also of [math]\displaystyle{ \,X^TX \, }[/math]), and they correspond to the columns of [math]\displaystyle{ \,U }[/math] (also of [math]\displaystyle{ \,V }[/math]).
We are interested in [math]\displaystyle{ \,U }[/math], whose [math]\displaystyle{ \,d }[/math] columns are the [math]\displaystyle{ \,d }[/math] directions of variation of our data. Ordered from left to right, the [math]\displaystyle{ \,ith }[/math] column of [math]\displaystyle{ \,U }[/math] is the [math]\displaystyle{ \,ith }[/math] most informative direction of variation of our data. That is, the [math]\displaystyle{ \,ith }[/math] column of [math]\displaystyle{ \,U }[/math] is the [math]\displaystyle{ \,ith }[/math] most effective column in terms of capturing the total variance exhibited by our data. A subset of the columns of [math]\displaystyle{ \,U }[/math] is used by PCA to reduce the dimensionality of [math]\displaystyle{ \,X }[/math] by projecting [math]\displaystyle{ \,X }[/math] onto the columns of this subset. In practice, when we apply PCA to [math]\displaystyle{ \,X }[/math] to reduce the dimensionality of [math]\displaystyle{ \,X }[/math] from [math]\displaystyle{ \,d }[/math] to [math]\displaystyle{ \,k }[/math], where [math]\displaystyle{ k \lt d\, }[/math], we would proceed as follows:
- Step 1: Center [math]\displaystyle{ \,X }[/math] so that it would have zero mean.
- Step 2: Apply singular value decomposition to [math]\displaystyle{ \,X }[/math] to obtain [math]\displaystyle{ \,U }[/math].
- Step 3: Suppose we denote the resulting [math]\displaystyle{ \,k }[/math]-dimensional representation of [math]\displaystyle{ \,X }[/math] by [math]\displaystyle{ \,Y }[/math]. Then, [math]\displaystyle{ \,Y }[/math] is obtained as [math]\displaystyle{ \,Y = U_k^TX }[/math]. Here, [math]\displaystyle{ \,U_k }[/math] consists of the first (leftmost) [math]\displaystyle{ \,k }[/math] columns of [math]\displaystyle{ \,U }[/math] that correspond to the [math]\displaystyle{ \,k }[/math] largest diagonal elements of [math]\displaystyle{ \,\Sigma }[/math].
PCA takes a sample of d - dimensional vectors and produces an orthogonal(zero covariance) set of d 'Principal Components'. The first Principal Component is the direction of greatest variance in the sample. The second principal component is the direction of second greatest variance (orthogonal to the first component), etc.
Then we can preserve most of the variance in the sample in a lower dimension by choosing the first k Principle Components and approximating the data in k - dimensional space, which is easier to analyze and plot.
Principal Components of handwritten digits
Suppose that we have a set of 130 images (28 by 23 pixels) of handwritten threes. {{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: This figure is copyrighted. Please remove and replace it with an appropriate one. You can produce an image yourself using 3 digits in 2-3 data set for example.. Please improve this article if you can. (September 6 2010) | small = | smallimage = | smallimageright = | smalltext = }} {{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: If anyone can tell me where I can find the 2-3 data set, I would create the new image. In the mean time, I found a non-copyrighted image of different looking 3s online, but as you can see, it is not as nice as one we could make.. Please improve this article if you can. (September 6 2010) | small = | smallimage = | smallimageright = | smalltext = }}
We can represent each image as a vector of length 644 ([math]\displaystyle{ 644 = 23 \times 28 }[/math]). Then we can represent the entire data set as a 644 by 130 matrix, shown below. Each column represents one image (644 rows = 644 pixels).
Using PCA, we can approximate the data as the product of two smaller matrices, which I will call [math]\displaystyle{ V \in M_{644,2} }[/math] and [math]\displaystyle{ W \in M_{2,103} }[/math]. If we expand the matrix product then each image is approximated by a linear combination of the columns of V: [math]\displaystyle{ \hat{f}(\lambda) = \bar{x} + \lambda_1 v_1 + \lambda_2 v_2 }[/math], where [math]\displaystyle{ \lambda = [\lambda_1, \lambda_2]^T }[/math] is a column of W.
To demonstrate this process, we can compare the images of 2s and 3s. We will apply PCA to the data, and compare the images of the labeled data. This is an example in classifying.
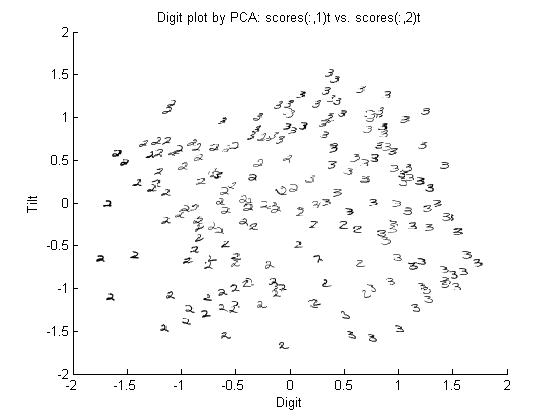
Don't worry about the constant term for now. The point is that we can represent an image using just 2 coefficients instead of 644. Also notice that the coefficients correspond to features of the handwritten digits. The picture below shows the first two principal components for the set of handwritten threes.
The first coefficient represents the width of the entire digit, and the second coefficient represents the slant of each handwritten digit.
Derivation of the first Principle Component
{{
Template:namespace detect
| type = style
| image =
| imageright =
| style =
| textstyle =
| text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: I think English of this section must be improved. Please improve this article if you can. (October 2010)
| small =
| smallimage =
| smallimageright =
| smalltext =
}}
We want to find the direction of maximum variation. Let [math]\displaystyle{ \begin{align}\textbf{w}\end{align} }[/math] be an arbitrary direction, [math]\displaystyle{ \begin{align}\textbf{x}\end{align} }[/math] a data point and [math]\displaystyle{ \begin{align}\displaystyle u\end{align} }[/math] the length of the projection of [math]\displaystyle{ \begin{align}\textbf{x}\end{align} }[/math] in direction [math]\displaystyle{ \begin{align}\textbf{w}\end{align} }[/math].
[math]\displaystyle{ \begin{align}
\textbf{w} &= [w_1, \ldots, w_D]^T \\
\textbf{x} &= [x_1, \ldots, x_D]^T \\
u &= \frac{\textbf{w}^T \textbf{x}}{\sqrt{\textbf{w}^T\textbf{w}}}
\end{align}
}[/math]
The direction [math]\displaystyle{ \begin{align}\textbf{w}\end{align} }[/math] is the same as [math]\displaystyle{ \begin{align}c\textbf{w}\end{align} }[/math], for any scalar [math]\displaystyle{ c }[/math], so without loss of generality,
[math]\displaystyle{
\begin{align}
|\textbf{w}| &= \sqrt{\textbf{w}^T\textbf{w}} = 1 \\
u &= \textbf{w}^T \textbf{x}.
\end{align}
}[/math]
Let [math]\displaystyle{ x_1, \ldots, x_D }[/math] be random variables, then our goal is to maximize the variance of [math]\displaystyle{ u }[/math],
[math]\displaystyle{
\textrm{var}(u) = \textrm{var}(\textbf{w}^T \textbf{x}) = \textbf{w}^T \Sigma \textbf{w}.
}[/math]
For a finite data set we replace the covariance matrix [math]\displaystyle{ \Sigma }[/math] by [math]\displaystyle{ s }[/math], the sample covariance matrix
[math]\displaystyle{ \textrm{var}(u) = \textbf{w}^T s\textbf{w} . }[/math]
The above is the variance of [math]\displaystyle{ \begin{align}\displaystyle u \end{align} }[/math] formed by the weight vector [math]\displaystyle{ \begin{align}\textbf{w} \end{align} }[/math]. The first principal component is the vector [math]\displaystyle{ \begin{align}\textbf{w} \end{align} }[/math] that maximizes the variance,
[math]\displaystyle{
\textrm{PC} = \underset{\textbf{w}}{\operatorname{arg\,max}} \, \left( \operatorname{var}(u) \right) = \underset{\textbf{w}}{\operatorname{arg\,max}} \, \left( \textbf{w}^T s \textbf{w} \right)
}[/math]
where arg max denotes the value of [math]\displaystyle{ \begin{align}\textbf{w} \end{align} }[/math] that maximizes the function. Our goal is to find the weight [math]\displaystyle{ \begin{align}\textbf{w} \end{align} }[/math] that maximizes this variability, subject to a constraint. Since our function is convex, it has no maximum value. Therefore we need to add a constraint that restricts the length of [math]\displaystyle{ \begin{align}\textbf{w} \end{align} }[/math]. However, we are only interested in the direction of the variability, so the problem becomes
[math]\displaystyle{
\underset{\textbf{w}}{\operatorname{max}} \, \left( \textbf{w}^T s \textbf{w} \right)
}[/math]
s.t. [math]\displaystyle{ \textbf{w}^T \textbf{w} = 1. }[/math]
Notice,
[math]\displaystyle{
\textbf{w}^T s \textbf{w} \leq \| \textbf{w}^T s \textbf{w} \| \leq \| s \| \| \textbf{w} \| = \| s \|.
}[/math]
Therefore the variance is bounded, so the maximum exists. We find the this maximum using the method of Lagrange multipliers.
Lagrange Multiplier
Before we can proceed, we must review Lagrange Multipliers.
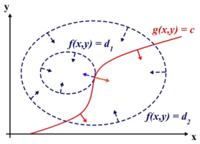
To find the maximum (or minimum) of a function [math]\displaystyle{ \displaystyle f(x,y) }[/math] subject to constraints [math]\displaystyle{ \displaystyle g(x,y) = 0 }[/math], we define a new variable [math]\displaystyle{ \displaystyle \lambda }[/math] called a Lagrange Multiplier and we form the Lagrangian,
[math]\displaystyle{ \displaystyle L(x,y,\lambda) = f(x,y) - \lambda g(x,y) }[/math]
If [math]\displaystyle{ \displaystyle (x^*,y^*) }[/math] is the max of [math]\displaystyle{ \displaystyle f(x,y) }[/math], there exists [math]\displaystyle{ \displaystyle \lambda^* }[/math] such that [math]\displaystyle{ \displaystyle (x^*,y^*,\lambda^*) }[/math] is a stationary point of [math]\displaystyle{ \displaystyle L }[/math] (partial derivatives are 0).
In addition [math]\displaystyle{ \displaystyle (x^*,y^*) }[/math] is a point in which functions [math]\displaystyle{ \displaystyle f }[/math] and [math]\displaystyle{ \displaystyle g }[/math] touch but do not cross. At this point, the tangents of [math]\displaystyle{ \displaystyle f }[/math] and [math]\displaystyle{ \displaystyle g }[/math] are parallel or gradients of [math]\displaystyle{ \displaystyle f }[/math] and [math]\displaystyle{ \displaystyle g }[/math] are parallel, such that:
[math]\displaystyle{ \displaystyle \nabla_{x,y } f = \lambda \nabla_{x,y } g }[/math]
where,
[math]\displaystyle{ \displaystyle \nabla_{x,y} f = (\frac{\partial f}{\partial x},\frac{\partial f}{\partial{y}}) \leftarrow }[/math] the gradient of [math]\displaystyle{ \, f }[/math]
[math]\displaystyle{ \displaystyle \nabla_{x,y} g = (\frac{\partial g}{\partial{x}},\frac{\partial{g}}{\partial{y}}) \leftarrow }[/math] the gradient of [math]\displaystyle{ \, g }[/math]
Example
Suppose we wish to maximise the function [math]\displaystyle{ \displaystyle f(x,y)=x-y }[/math] subject to the constraint [math]\displaystyle{ \displaystyle x^{2}+y^{2}=1 }[/math]. We can apply the Lagrange multiplier method on this example; the lagrangian is:
[math]\displaystyle{ \displaystyle L(x,y,\lambda) = x-y - \lambda (x^{2}+y^{2}-1) }[/math]
We want the partial derivatives equal to zero:
[math]\displaystyle{ \displaystyle \frac{\partial L}{\partial x}=1+2 \lambda x=0 }[/math]
[math]\displaystyle{ \displaystyle \frac{\partial L}{\partial y}=-1+2\lambda y=0 }[/math]
[math]\displaystyle{ \displaystyle \frac{\partial L}{\partial \lambda}=x^2+y^2-1 }[/math]
Solving the system we obtain 2 stationary points: [math]\displaystyle{ \displaystyle (\sqrt{2}/2,-\sqrt{2}/2) }[/math] and [math]\displaystyle{ \displaystyle (-\sqrt{2}/2,\sqrt{2}/2) }[/math]. In order to understand which one is the maximum, we just need to substitute it in [math]\displaystyle{ \displaystyle f(x,y) }[/math] and see which one as the biggest value. In this case the maximum is [math]\displaystyle{ \displaystyle (\sqrt{2}/2,-\sqrt{2}/2) }[/math].
Determining W
Back to the original problem, from the Lagrangian we obtain,
[math]\displaystyle{ \displaystyle L(\textbf{w},\lambda) = \textbf{w}^T S \textbf{w} - \lambda (\textbf{w}^T \textbf{w} - 1) }[/math]
If [math]\displaystyle{ \textbf{w}^T \textbf{w} }[/math] is a unit vector then the second part of the equation is 0.
If [math]\displaystyle{ \textbf{w}^T \textbf{w} }[/math] is not a unit vector then the second part of the equation increases. Thus decreasing overall [math]\displaystyle{ \displaystyle L(\textbf{w},\lambda) }[/math]. Maximization happens when [math]\displaystyle{ \textbf{w}^T \textbf{w} =1 }[/math]
(Note that to take the derivative with respect to w below, [math]\displaystyle{ \textbf{w}^T S \textbf{w} }[/math] can be thought of as a quadratic function of w, hence the 2sw below. For more matrix derivatives, see section 2 of the Matrix Cookbook)
Taking the derivative with respect to w, we get:
[math]\displaystyle{ \displaystyle \frac{\partial L}{\partial \textbf{w}} = 2S\textbf{w} - 2\lambda\textbf{w} }[/math]
Set [math]\displaystyle{ \displaystyle \frac{\partial L}{\partial \textbf{w}} = 0 }[/math], we get
[math]\displaystyle{ \displaystyle S\textbf{w}^* = \lambda^*\textbf{w}^* }[/math]
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: It is good discussion, what will happen if we don't have distinct eigenvalues and eigenvectors? What does this situation mean?. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }} {{
Template:namespace detect
| type = style
| image =
| imageright =
| style =
| textstyle =
| text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: If the eigenvalues are not distinct, I suppose we could still take the leftmost eigenvector by default. Not sure if this is the correct approach, so can anyone please explain further? Thanks. Please improve this article if you can. (October 2010)
| small =
| smallimage =
| smallimageright =
| smalltext =
}}
From the eigenvalue equation [math]\displaystyle{ \, \textbf{w}^* }[/math] is an eigenvector of S and [math]\displaystyle{ \, \lambda^* }[/math] is the corresponding eigenvalue of S. If we substitute [math]\displaystyle{ \displaystyle\textbf{w}^* }[/math] in [math]\displaystyle{ \displaystyle \textbf{w}^T S\textbf{w} }[/math] we obtain,
[math]\displaystyle{ \displaystyle\textbf{w}^{*T} S\textbf{w}^* = \textbf{w}^{*T} \lambda^* \textbf{w}^* = \lambda^* \textbf{w}^{*T} \textbf{w}^* = \lambda^* }[/math]
In order to maximize the objective function we choose the eigenvector corresponding to the largest eigenvalue. We choose the first PC, u1 to have the maximum variance
(i.e. capturing as much variability in [math]\displaystyle{ \displaystyle x_1, x_2,...,x_D }[/math] as possible.) Subsequent principal components will take up successively smaller parts of the total variability.
D dimensional data will have D eigenvectors
[math]\displaystyle{ \lambda_1 \geq \lambda_2 \geq ... \geq \lambda_D }[/math] where each [math]\displaystyle{ \, \lambda_i }[/math] represents the amount of variation in direction [math]\displaystyle{ \, i }[/math]
so that
[math]\displaystyle{ Var(u_1) \geq Var(u_2) \geq ... \geq Var(u_D) }[/math]
Note that the Principal Components decompose the total variance in the data:
[math]\displaystyle{ \displaystyle \sum_{i = 1}^D Var(u_i) = \sum_{i = 1}^D \lambda_i = Tr(S) = Var(\sum_{i = 1}^n x_i) }[/math]
i.e. the sum of variations in all directions is the variation in the whole data
Example from class
We apply PCA to the noise data, making the assumption that the intrinsic dimensionality of the data is 10. We now try to compute the reconstructed images using the top 10 eigenvectors and plot the original and reconstructed images
The Matlab code is as follows:
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: I think as mentioned in the class this code does not perform PCA, since mean of the source vector has not been subtracted. This code mus be altered so as to implement real PCA. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }}
load noisy who size(X) imagesc(reshape(X(:,1),20,28)') colormap gray imagesc(reshape(X(:,1),20,28)') [u s v] = svd(X); xHat = u(:,1:10)*s(1:10,1:10)*v(:,1:10)'; % use ten principal components figure imagesc(reshape(xHat(:,1000),20,28)') % here '1000' can be changed to different values, e.g. 105, 500, etc. colormap gray
Running the above code gives us 2 images - the first one represents the noisy data - we can barely make out the face
The second one is the denoised image
-
"Noisy Face"
-
"De-noised Face"
As you can clearly see, more features can be distinguished from the picture of the de-noised face compared to the picture of the noisy face. This is because almost all of the noise in the noisy image is captured by the principal components (directions of variation) that capture the least amount of variation in the image, and these principal components were discarded when we used the few principal components that capture most of the image's variation to generate the image's lower-dimensional representation. If we took more principal components, at first the image would improve since the intrinsic dimensionality is probably more than 10. But if you include all the components you get the noisy image, so not all of the principal components improve the image. In general, it is difficult to choose the optimal number of components.
Application of PCA - Feature Extraction
One of the applications of PCA is to group similar data (e.g. images). There are generally two methods to do this. We can classify the data (i.e. give each data a label and compare different types of data) or cluster (i.e. do not label the data and compare output for classes).
Generally speaking, we can do this with the entire data set (if we have an 8X8 picture, we can use all 64 pixels). However, this is hard, and it is easier to use the reduced data and features of the data.
General PCA Algorithm
The PCA Algorithm is summarized as follows (taken from the Lecture Slides).
Algorithm
Recover basis: Calculate [math]\displaystyle{ XX^T =\Sigma_{i=1}^{n} x_i x_{i}^{T} }[/math] and let [math]\displaystyle{ U= }[/math] eigenvectors of [math]\displaystyle{ X X^T }[/math] corresponding to the top [math]\displaystyle{ d }[/math] eigenvalues.
Encoding training data: Let [math]\displaystyle{ Y=U^TX }[/math] where [math]\displaystyle{ Y }[/math] is a [math]\displaystyle{ d \times n }[/math] matrix of encoding of the original data.
Reconstructing training data: [math]\displaystyle{ \hat{X}= UY=UU^TX }[/math].
Encode set example: [math]\displaystyle{ y=U^T x }[/math] where [math]\displaystyle{ y }[/math] is a [math]\displaystyle{ d- }[/math]dimentional encoding of [math]\displaystyle{ x }[/math].
Reconstruct test example: [math]\displaystyle{ \hat{x}= Uy=UU^Tx }[/math].
Other Notes:
- The mean of the data(X) must be 0. This means we may have to preprocess the data by subtracting off the mean(see detailsPCA in Wikipedia.)
- Encoding the data means that we are projecting the data onto a lower dimensional subspace by taking the inner product. Encoding: [math]\displaystyle{ X_{D\times n} \longrightarrow Y_{d\times n} }[/math] using mapping [math]\displaystyle{ \, U^T X_{D \times n} }[/math].
- When we reconstruct the training set, we are only using the top d dimensions.This will eliminate the dimensions that have lower variance (e.g. noise). Reconstructing: [math]\displaystyle{ \hat{X}_{D\times n}\longleftarrow Y_{d \times n} }[/math] using mapping [math]\displaystyle{ \, U_dY_{d \times n} }[/math], where [math]\displaystyle{ \,U_d }[/math] contains the first (leftmost) [math]\displaystyle{ \,d }[/math] columns of [math]\displaystyle{ \,U }[/math].
- We can compare the reconstructed test sample to the reconstructed training sample to classify the new data.
Fisher's (Linear) Discriminant Analysis (FDA) - Two Class Problem - October 5, 2010
Sir Ronald A. Fisher
Fisher's Discriminant Analysis (FDA), also known as Fisher's Linear Discriminant Analysis (LDA) in some sources, is a classical feature extraction technique. It was originally described in 1936 by Sir Ronald Aylmer Fisher, an English statistician and eugenicist who has been described as one of the founders of modern statistical science. His original paper describing FDA can be found here; a Wikipedia article summarizing the algorithm can be found here. In this paper Fisher used for the first time the term DISCRIMINANT FUNCTION. The term DISCRIMINANT ANALYSIS was introduced later by Fisher himself in a subsequent paper which can be found here.
Contrasting FDA with PCA
Similar to PCA, the goal of FDA is to project the data in a lower dimension. The main difference between FDA and PCA is that, in FDA, in contrast to PCA, we are not interested in retaining as much of the variance of our original data as possible. Rather, in FDA, our goal is to find a direction that is useful for classifying the data (i.e. in this case, we are looking for a direction that is most representative of a particular characteristic e.g. glasses vs. no-glasses). Suppose we have 2-dimensional data, then FDA would attempt to project the data of each class onto a point in such a way that the resulting two points would be as far apart from each other as possible. Intuitively, this basic idea behind FDA is the optimal way for separating each pair of classes along a certain direction.
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: Just a thought: how relevant is "Dimensionality reduction techniques" to the concept of "subspace clustering"? As in subspace clustering, the goal is to find a set of features (relevant features, the concept is referred to as local feature relevance in the literature) in the high dimensional space, where potential subspaces accommodating different classes of data points can be defined. This means; the data points are dense when they are considered in a subset of dimensions (features).. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }} {{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: If I'm not mistaken, classification techniques like FDA use labeled training data whereas clustering techniques use unlabeled training data instead. Any other input regarding this would be much appreciated. Thanks. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }}
The number of dimensions that we want to reduce the data to depends on the number of classes:
For a 2-classes problem, we want to reduce the data to one dimension (a line), [math]\displaystyle{ \displaystyle Z \in \mathbb{R}^{1} }[/math]
Generally, for a k-classes problem, we want to reduce the data to k-1 dimensions, [math]\displaystyle{ \displaystyle Z \in \mathbb{R}^{k-1} }[/math]
As we will see from our objective function, we want to maximize the separation of the classes and to minimize the within-variance of each class. That is, our ideal situation is that the individual classes are as far away from each other as possible, and at the same time the data within each class are as close to each other as possible (collapsed to a single point in the most extreme case).
The following diagram summarizes this goal.
In fact, the two examples above may represent the same data projected on two different lines.
Distance Metric Learning VS FDA
In many fundamental machine learning problems, the Euclidean distances between data points do not represent the desired topology that we are trying to capture. Kernel methods address this problem by mapping the points into new spaces where Euclidean distances may be more useful. An alternative approach is to construct a Mahalanobis distance (quadratic Gaussian metric) over the input space and use it in place of Euclidean distances. This approach can be equivalently interpreted as a linear transformation of the original inputs, followed by Euclidean distance in the projected space. This approach has attracted a lot of recent interest.
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: Anyone please add an example to make the comparison clearer. Please improve this article if you can. (October2010) | small = | smallimage = | smallimageright = | smalltext = }}
Some of the proposed algorithms are iterative and computationally expensive. In the paper,"Distance Metric Learning VS FDA " written by our instructor, they propose a closed-form solution to one algorithm that previously required expensive semidefinite optimization. They provide a new problem setup in which the algorithm performs better or as well as some standard methods, but without the computational complexity. Furthermore, they show a strong relationship between these methods and the Fisher Discriminant Analysis (FDA). They also extend the approach by kernelizing it, allowing for non-linear transformations of the metric.
FDA Goals
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: It would be nice to see how did Fisher arrived at these two goals. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }} FDA projects the data into lower dimensional space, where the distances between the projected means are maximum and the within-class variances are minimum. There are two categories of classification problems:
1. Two-class problem
2. Multi-class problem (addressed next lecture)
Two-class problem
In the two-class problem, we have the pre-knowledge that data points belong to two classes. Intuitively speaking points of each class form a cloud around the mean of the class, with each class having possibly different size. To be able to separate the two classes we must determine the class whose mean is closest to a given point while also accounting for the different size of each class, which is represented by the covariance of each class.
Assume [math]\displaystyle{ \underline{\mu_{1}}=\frac{1}{n_{1}}\displaystyle\sum_{i:y_{i}=1}\underline{x_{i}} }[/math] and [math]\displaystyle{ \displaystyle\Sigma_{1} }[/math], represent the mean and covariance of the 1st class, and [math]\displaystyle{ \underline{\mu_{2}}=\frac{1}{n_{2}}\displaystyle\sum_{i:y_{i}=2}\underline{x_{i}} }[/math] and [math]\displaystyle{ \displaystyle\Sigma_{2} }[/math] represent the mean and covariance of the 2nd class. We have to find a transformation which satisfies the following goals:
1.To make the means of these two classes as far apart as possible
- In other words, the goal is to maximize the distance after projection between class 1 and class 2. This can be done by maximizing the distance between the means of the classes after projection. When projecting the data points to a one-dimensional space, all points will be projected to a single line; the line we seek is the one with the direction that achieves maximum separation of classes upon projection. If the original points are [math]\displaystyle{ \underline{x_{i}} \in \mathbb{R}^{d} }[/math]and the projected points are [math]\displaystyle{ \underline{w}^T \underline{x_{i}} }[/math] then the mean of the projected points will be [math]\displaystyle{ \underline{w}^T \underline{\mu_{1}} }[/math] and [math]\displaystyle{ \underline{w}^T \underline{\mu_{2}} }[/math] for class 1 and class 2 respectively. The goal now becomes to maximize the Euclidean distance between projected means, [math]\displaystyle{ (\underline{w}^T\underline{\mu_{1}}-\underline{w}^T\underline{\mu_{2}})^T (\underline{w}^T\underline{\mu_{1}}-\underline{w}^T\underline{\mu_{2}}) }[/math]. The steps of this maximization are given below.
2.We want to collapse all data points of each class to a single point, i.e., minimize the covariance within each class
- Notice that the variance of the projected classes 1 and 2 are given by [math]\displaystyle{ \underline{w}^T\Sigma_{1}\underline{w} }[/math] and [math]\displaystyle{ \underline{w}^T\Sigma_{2}\underline{w} }[/math]. The second goal is to minimize the sum of these two covariances (the summation of the two covariances is a valid covariance, satisfying the symmetry and positive semi-definite criteria).
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: In 2. above, I wonder if the computation would be much more complex if we instead find a weighted sum of the covariances of the two classes where the weights are the sizes of the two classes?. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }}
As is demonstrated below, both of these goals can be accomplished simultaneously.
Original points are [math]\displaystyle{ \underline{x_{i}} \in \mathbb{R}^{d} }[/math]
[math]\displaystyle{ \ \{ \underline x_1 \underline x_2 \cdot \cdot \cdot \underline x_n \} }[/math]
Projected points are [math]\displaystyle{ \underline{z_{i}} \in \mathbb{R}^{1} }[/math] with [math]\displaystyle{ \underline{z_{i}} = \underline{w}^T \cdot\underline{x_{i}} }[/math] where [math]\displaystyle{ \ z_i }[/math] is a scalar
1. Minimizing within-class variance
[math]\displaystyle{ \displaystyle \min_w (\underline{w}^T\sum_1\underline{w}) }[/math]
[math]\displaystyle{ \displaystyle \min_w (\underline{w}^T\sum_2\underline{w}) }[/math]
and this problem reduces to [math]\displaystyle{ \displaystyle \min_w (\underline{w}^T(\sum_1 + \sum_2)\underline{w}) }[/math]
(where [math]\displaystyle{ \,\sum_1 }[/math] and [math]\displaystyle{ \,\sum_2 }[/math] are the covariance matrices of the 1st and 2nd classes of data, respectively)
Let [math]\displaystyle{ \displaystyle \ s_w=\sum_1 + \sum_2 }[/math] be the within-classes covariance. Then, this problem can be rewritten as [math]\displaystyle{ \displaystyle \min_w (\underline{w}^Ts_w\underline{w}) }[/math].
2. Maximize the distance between the means of the projected data
[math]\displaystyle{ \displaystyle \max_w ||\underline{w}^T \mu_1 - \underline{w}^T \mu_2||^2, }[/math]
[math]\displaystyle{ \begin{align} ||\underline{w}^T \mu_1 - \underline{w}^T \mu_2||^2 &= (\underline{w}^T \mu_1 - \underline{w}^T \mu_2)^T(\underline{w}^T \mu_1 - \underline{w}^T \mu_2)\\
&= (\mu_1^T\underline{w} - \mu_2^T\underline{w})(\underline{w}^T \mu_1 - \underline{w}^T \mu_2)\\
&= (\mu_1 - \mu_2)^T \underline{w} \underline{w}^T (\mu_1 - \mu_2) \\
&= ((\mu_1 - \mu_2)^T \underline{w})^{T} (\underline{w}^T (\mu_1 - \mu_2))^{T} \\
&= \underline{w}^T(\mu_1 - \mu_2)(\mu_1 - \mu_2)^T \underline{w} \end{align} }[/math]
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: Matrix multiplication is in general not commutative. Therefore, in the last line in the derivation above, I wonder if this is always true? Since w is a vector multiplying the transpose with the difference between the means is a scalar. Therefore we can take the transpose and not change anything. Add [math]\displaystyle{ &= ((\mu_1 - \mu_2)^T \underline{w})^{T} (\underline{w}^T (\mu_1 - \mu_2))^{T} \\ }[/math] before the last line and the derivation follows from there.. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }}
Let [math]\displaystyle{ \displaystyle s_B = (\mu_1 - \mu_2)(\mu_1 - \mu_2)^T }[/math], the between-class covariance, then the goal is to [math]\displaystyle{ \displaystyle \max_w (\underline{w}^T(\mu_1 - \mu_2)(\mu_1 - \mu_2)^T\underline{w}) }[/math] or [math]\displaystyle{ \displaystyle \max_w (\underline{w}^Ts_B\underline{w}) }[/math].
The Objective Function for FDA
We want an objective function which satisfies both of the goals outlined above (at the same time).
- [math]\displaystyle{ \displaystyle \min_w (\underline{w}^T(\sum_1 + \sum_2)\underline{w}) }[/math] or [math]\displaystyle{ \displaystyle \min_w (\underline{w}^Ts_w\underline{w}) }[/math]
- [math]\displaystyle{ \displaystyle \max_w (\underline{w}^T(\mu_1 - \mu_2)(\mu_1 - \mu_2)^T\underline{w}) }[/math] or [math]\displaystyle{ \displaystyle \max_w (\underline{w}^Ts_B\underline{w}) }[/math]
So, we construct our objective function as maximizing the ratio of the two goals brought above:
[math]\displaystyle{ \displaystyle \max_w \frac{(\underline{w}^T(\mu_1 - \mu_2)(\mu_1 - \mu_2)^T\underline{w})} {(\underline{w}^T(\sum_1 + \sum_2)\underline{w})} }[/math]
or equivalently,
[math]\displaystyle{ \displaystyle \max_w \frac{(\underline{w}^Ts_B\underline{w})}{(\underline{w}^Ts_w\underline{w})} }[/math]
One may argue that we can use subtraction for this purpose, while this approach is true but it can be shown it will need another scaling factor. Thus using this ratio is more efficient.
In order to solve this optimization problem, we form the Lagrangian:
[math]\displaystyle{ \displaystyle L(\underline{w},\lambda) = \underline{w}^Ts_B\underline{w} - \lambda (\underline{w}^Ts_w\underline{w} -1) }[/math]
Then, we equate the partial derivative of L w.r.to [math]\displaystyle{ \underline{w} }[/math]:
[math]\displaystyle{ \displaystyle \frac{\partial L}{\partial \underline{w}}=2s_B \underline{w} - 2\lambda s_w \underline{w} = 0 }[/math]
[math]\displaystyle{ s_B \underline{w} = \lambda s_w \underline{w} }[/math]
[math]\displaystyle{ s_w^{-1}s_B \underline{w}= \lambda\underline{w} }[/math]
This is in the form of generalized eigenvalue problem. Therefore, [math]\displaystyle{ \underline{w} }[/math] is the largest eigenvector of [math]\displaystyle{ s_w^{-1}s_B }[/math]
This solution can be further simplified as follow:
[math]\displaystyle{ s_w^{-1}(\mu_1 - \mu_2)(\mu_1 - \mu_2)^T\underline{w} = \lambda\underline{w} }[/math]
Since [math]\displaystyle{ (\mu_1 - \mu_2)^T\underline{w} }[/math] is a scalar then [math]\displaystyle{ s_w^{-1}(\mu_1 - \mu_2) }[/math]∝[math]\displaystyle{ \underline{w} }[/math]
This gives the direction of [math]\displaystyle{ \underline{w} }[/math] without doing eigenvalue decomposition in the case of 2-class problem.
Note: In order for [math]\displaystyle{ {s_w} }[/math] to have an inverse, it must have full rank. This can be achieved by ensuring that the number of data points >= dimensionality of [math]\displaystyle{ \underline{x_{i}} }[/math].
FDA Using Matlab
Note: The following example was not actually mentioned in this lecture
We see now an application of the theory that we just introduced. Using Matlab, we find the principal components and the projection by Fisher Discriminant Analysis of two Bivariate normal distributions to show the difference between the two methods.
%First of all, we generate the two data set:
% First data set X1
X1 = mvnrnd([1,1],[1 1.5; 1.5 3], 300);
%In this case:
mu_1=[1;1];
Sigma_1=[1 1.5; 1.5 3];
%where mu and sigma are the mean and covariance matrix.
% Second data set X2
X2 = mvnrnd([5,3],[1 1.5; 1.5 3], 300);
%Here mu_2=[5;3] and Sigma_2=[1 1.5; 1.5 3]
%The plot of the two distributions is:
plot(X1(:,1),X1(:,2),'.b'); hold on;
plot(X2(:,1),X2(:,2),'ob')
%We compute the principal components:
% Combine data sets to map both into the same subspace
X=[X1;X2];
X=X';
% We used built-in PCA function in Matlab
[coefs, scores]=princomp(X);
plot([0 coefs(1,1)], [0 coefs(2,1)],'b')
plot([0 coefs(1,1)]*10, [0 coefs(2,1)]*10,'r')
sw=2*[1 1.5;1.5 3] % sw=Sigma1+Sigma2=2*Sigma1
w=sw\[4; 2] % calculate s_w^{-1}(mu2 - mu1)
plot ([0 w(1)], [0 w(2)],'g')
%We now make the projection:
Xf=w'*X
figure
plot(Xf(1:300),1,'ob') %In this case, since it's a one dimension data, the plot is "Data Vs Indexes"
hold on
plot(Xf(301:600),1,'or')
%We see that in the above picture that there is no overlapping
Xp=coefs(:,1)'*X
figure
plot(Xp(1:300),1,'b')
hold on
plot(Xp(301:600),2,'or')
%In this case there is an overlapping since we project the first principal component on [Xp=coefs(:,1)'*X]
Some of FDA applications
There are many applications for FDA in many domains some of them are stated below:
- SPEECH/MUSIC/NOISE CLASSIFICATION IN HEARING AIDS
FDA can be used to enhance listening comprehension when the user goes from a sound environment to another different one. For more information review this paper by Alexandre et al.here
- Application to Face Recognition
FDA can be used in face recognition at different situation. Using FDA Kong et al. proposes an Application to Face Recognition with Small Number of Training Samples here.
- Palmprint Recognition
FDA is used in biometrics, to implement an automated palmprint recognition system. See An Automated Palmprint Recognition System by Tee et al. here.
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: I think briefing about the other applications would be easier than browsing through all of these applications. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }}
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: This link is no longer valid.. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }}
other applications could found in references 4,5,6,7,8 and more in here
References
1. Kong, H.; Wang, L.; Teoh, E.K.; Wang, J.-G.; Venkateswarlu, R.; , "A framework of 2D Fisher discriminant analysis: application to face recognition with small number of training samples," Computer Vision and Pattern Recognition, 2005. CVPR 2005. IEEE Computer Society Conference on , vol.2, no., pp. 1083- 1088 vol. 2, 20-25 June 2005 doi: 10.1109/CVPR.2005.30 1
2. Enrique Alexandre, Roberto Gil-Pita, Lucas Cuadra, Lorena A´lvarez, Manuel Rosa-Zurera, "SPEECH/MUSIC/NOISE CLASSIFICATION IN HEARING AIDS USING A TWO-LAYER CLASSIFICATION SYSTEM WITH MSE LINEAR DISCRIMINANTS", 16th European Signal Processing Conference (EUSIPCO 2008), Lausanne, Switzerland, August 25-29, 2008, copyright by EURASIP, 2
3. Connie, Tee; Jin, Andrew Teoh Beng; Ong, Michael Goh Kah; Ling, David Ngo Chek; "An automated palmprint recognition system", Journal of Image and Vision Computing, 2005. 3
4. met, Francesca; Boqué, Ricard; Ferré, Joan; "Application of non-negative matrix factorization combined with Fisher's linear discriminant analysis for classification of olive oil excitation-emission fluorescence spectra", Journal of Chemometrics and Intelligent Laboratory Systems, 2006. 4
5. Chiang, Leo H.;Kotanchek, Mark E.;Kordon, Arthur K.; "Fault diagnosis based on Fisher discriminant analysis and support vector machines" Journal of Computers & Chemical Engineering, 2004 5
6. Yang, Jian ;Frangi, Alejandro F.; Yang, Jing-yu; "A new kernel Fisher discriminant algorithm with application to face recognition", 2004 6
7. Cawley, Gavin C.; Talbot, Nicola L. C.; "Efficient leave-one-out cross-validation of kernel fisher discriminant classifiers", Journal of Pattern Recognition , 2003 7
8. Kodipaka, S.; Vemuri, B.C.; Rangarajan, A.; Leonard, C.M.; Schmallfuss, I.; Eisenschenk, S.; "Kernel Fisher discriminant for shape-based classification in epilepsy" Hournal Medical Image Analysis, 2007. 8
Fisher's (Linear) Discriminant Analysis (FDA) - Multi-Class Problem - October 7, 2010
Obtaining Covariance Matrices
The within-class covariance matrix [math]\displaystyle{ \mathbf{S}_{W} }[/math] is easy to obtain:
- [math]\displaystyle{ \begin{align} \mathbf{S}_{W} = \sum_{i=1}^{k} \mathbf{S}_{W,i} \end{align} }[/math]
where [math]\displaystyle{ \mathbf{S}_{W,i} = \frac{1}{n_{i}}\sum_{j: y_{j}=i}(\mathbf{x}_{j} - \mathbf{\mu}_{i})(\mathbf{x}_{j} - \mathbf{\mu}_{i})^{T} }[/math] and [math]\displaystyle{ \mathbf{\mu}_{i} = \frac{\sum_{j: y_{j}=i}\mathbf{x}_{j}}{n_{i}} }[/math].
However, the between-class covariance matrix [math]\displaystyle{ \mathbf{S}_{B} }[/math] is not easy to compute directly. To bypass this problem we use the following method. We know that the total variance is constant, and so we decompose the variance into two parts: within-class and between-class (similar to ANOVA). We have:
- [math]\displaystyle{ \begin{align} \mathbf{S}_{T} = \mathbf{S}_{B} + \mathbf{S}_{W} \end{align} }[/math]
where the total variance is given by
- [math]\displaystyle{ \begin{align} \mathbf{S}_{T} = \frac{1}{n} \sum_{i}(\mathbf{x_{i}-\mu})(\mathbf{x_{i}-\mu})^{T} \end{align} }[/math]
We can now get [math]\displaystyle{ \mathbf{S}_{B} }[/math] from the relationship:
- [math]\displaystyle{ \begin{align} \mathbf{S}_{B} = \mathbf{S}_{T} - \mathbf{S}_{W} \end{align} }[/math]
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: Please check the derivation of decomposition of variance for errors. The total variance is missing a factor of 1/n, does this effect the formula for [math]\displaystyle{ \mathbf{S}_{B} }[/math]?. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }} {{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: You will still get the right results even without adding 1/n. However, the classes will be as if mirrored. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }}
Actually, there is another generation for [math]\displaystyle{ \mathbf{S}_{B} }[/math]. Denote a
total mean vector [math]\displaystyle{ \mathbf{\mu} }[/math] by
- [math]\displaystyle{ \begin{align} \mathbf{\mu} = \frac{1}{n}\sum_{i}\mathbf{x_{i}} = \frac{1}{n}\sum_{j=1}^{k}n_{j}\mathbf{\mu}_{j} \end{align} }[/math]
Thus the total covariance matrix [math]\displaystyle{ \mathbf{S}_{T} }[/math] is
- [math]\displaystyle{ \begin{align} \mathbf{S}_{T} = \sum_{i}(\mathbf{x_{i}-\mu})(\mathbf{x_{i}-\mu})^{T} \end{align} }[/math]
Thus we obtain
- [math]\displaystyle{ \begin{align} & \mathbf{S}_{T} = \sum_{i=1}^{k}\sum_{j: y_{j}=i}(\mathbf{x}_{j} - \mathbf{\mu}_{i} + \mathbf{\mu}_{i} - \mathbf{\mu})(\mathbf{x}_{j} - \mathbf{\mu}_{i} + \mathbf{\mu}_{i} - \mathbf{\mu})^{T} \\& = \sum_{i=1}^{k}\sum_{j: y_{j}=i}(\mathbf{x}_{j}-\mathbf{\mu}_{i})(\mathbf{x}_{j}-\mathbf{\mu}_{i})^{T}+ \sum_{i=1}^{k}\sum_{j: y_{j}=i}(\mathbf{\mu}_{i}-\mathbf{\mu})(\mathbf{\mu}_{i}-\mathbf{\mu})^{T} \\& = \mathbf{S}_{W} + \sum_{i=1}^{k} n_{i}(\mathbf{\mu}_{i}-\mathbf{\mu})(\mathbf{\mu}_{i}-\mathbf{\mu})^{T} \end{align} }[/math]
Since the total covariance [math]\displaystyle{ \mathbf{S}_{T} }[/math] is the sum of the within class covariance [math]\displaystyle{ \mathbf{S}_{W} }[/math] and the between class covariance [math]\displaystyle{ \mathbf{S}_{B} }[/math], we can denote the second term as the general between class covariance matrix [math]\displaystyle{ \mathbf{S}_{B} }[/math], thus we obtain
- [math]\displaystyle{ \begin{align} \mathbf{S}_{B} = \sum_{i=1}^{k} n_{i}(\mathbf{\mu}_{i}-\mathbf{\mu})(\mathbf{\mu}_{i}-\mathbf{\mu})^{T} \end{align} }[/math]
Therefore,
- [math]\displaystyle{ \begin{align} \mathbf{S}_{T} = \mathbf{S}_{W} + \mathbf{S}_{B} \end{align} }[/math]
Recall that in the two class case problem, we have
- [math]\displaystyle{ \begin{align} & \mathbf{S}_{B^{\ast}} = (\mathbf{\mu}_{1}-\mathbf{\mu}_{2})(\mathbf{\mu}_{1}-\mathbf{\mu}_{2})^{T} \\ & = (\mathbf{\mu}_{1}-\mathbf{\mu}+\mathbf{\mu}-\mathbf{\mu}_{2})(\mathbf{\mu}_{1}-\mathbf{\mu}+\mathbf{\mu}-\mathbf{\mu}_{2})^{T} \\ & = ((\mathbf{\mu}_{1}-\mathbf{\mu})-(\mathbf{\mu}_{2}-\mathbf{\mu}))((\mathbf{\mu}_{1}-\mathbf{\mu})-(\mathbf{\mu}_{2}-\mathbf{\mu}))^{T} \\ & = (\mathbf{\mu}_{1}-\mathbf{\mu})(\mathbf{\mu}_{1}-\mathbf{\mu})^{T}+(\mathbf{\mu}_{2}-\mathbf{\mu})(\mathbf{\mu}_{2}-\mathbf{\mu})^{T} \end{align} }[/math]
From the general form,
- [math]\displaystyle{ \begin{align} & \mathbf{S}_{B} = n_{1}(\mathbf{\mu}_{1}-\mathbf{\mu})(\mathbf{\mu}_{1}-\mathbf{\mu})^{T} + n_{2}(\mathbf{\mu}_{2}-\mathbf{\mu})(\mathbf{\mu}_{2}-\mathbf{\mu})^{T} \end{align} }[/math]
Apparently, they are very similar.
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: Please confirm that the algebra for the calculation of [math]\displaystyle{ \mathbf{S}_{B}^{\ast} }[/math] and [math]\displaystyle{ \mathbf{S}_{W}^{\ast} }[/math] is correct. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }} {{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: I Think it is not correct here are two terms missing with no explanations of why!!!!. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }}
Now, we are trying to find the optimal transformation. Basically, we have
- [math]\displaystyle{ \begin{align} \mathbf{z}_{i} = \mathbf{W}^{T}\mathbf{x}_{i}, i=1,2,...,k-1 \end{align} }[/math]
where [math]\displaystyle{ \mathbf{z}_{i} }[/math] is a [math]\displaystyle{ (k-1)\times 1 }[/math] vector, [math]\displaystyle{ \mathbf{W} }[/math] is a [math]\displaystyle{ d\times (k-1) }[/math] transformation matrix, i.e. [math]\displaystyle{ \mathbf{W} = [\mathbf{w}_{1}, \mathbf{w}_{2},..., \mathbf{w}_{k-1}] }[/math], and [math]\displaystyle{ \mathbf{x}_{i} }[/math] is a [math]\displaystyle{ d\times 1 }[/math] column vector.
Thus we obtain
- [math]\displaystyle{ \begin{align} & \mathbf{S}_{W}^{\ast} = \sum_{i=1}^{k}\sum_{j: y_{j}=i}(\mathbf{W}^{T}\mathbf{x}_{j}-\mathbf{W}^{T}\mathbf{\mu}_{i})(\mathbf{W}^{T}\mathbf{x}_{j}-\mathbf{W}^{T}\mathbf{\mu}_{i})^{T} \\ & = \sum_{i=1}^{k}\sum_{j: y_{j}=i}\mathbf{W}^{T}(\mathbf{x}_{j}-\mathbf{\mu}_{i})(\mathbf{x}_{j}-\mathbf{\mu}_{i})\mathbf{W} \\ & = \mathbf{W}^{T}\left[\sum_{i=1}^{k}\sum_{j: y_{j}=i}(\mathbf{x}_{j}-\mathbf{\mu}_{i})(\mathbf{x}_{j}-\mathbf{\mu}_{i})\right]\mathbf{W} \\ & = \mathbf{W}^{T}\mathbf{S}_{W}\mathbf{W} \end{align} }[/math]
Similarly, we obtain
- [math]\displaystyle{ \begin{align} & \mathbf{S}_{B}^{\ast} = \sum_{i=1}^{k}n_{i}(\mathbf{W}^{T}\mathbf{\mu}_{i}-\mathbf{W}^{T}\mathbf{\mu})(\mathbf{W}^{T}\mathbf{\mu}_{i}-\mathbf{W}^{T}\mathbf{\mu})^{T} \\ & = \sum_{i=1}^{k}n_{i}\mathbf{W}^{T}(\mathbf{\mu}_{i}-\mathbf{\mu})(\mathbf{\mu}_{i}-\mathbf{\mu})^{T}\mathbf{W} \\ & = \mathbf{W}^{T}\left[ \sum_{i=1}^{k}n_{i}(\mathbf{\mu}_{i}-\mathbf{\mu})(\mathbf{\mu}_{i}-\mathbf{\mu})^{T}\right]\mathbf{W} \\ & = \mathbf{W}^{T}\mathbf{S}_{B}\mathbf{W} \end{align} }[/math]
Now, we use the determinant of the matrix, i.e. the product of the eigenvalues of the matrix, as our measure.
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: There is no justification for using determinant. Moreover there is inconsistency here. Should we use Trace (as suggested below) or Determinant (as suggested here). Please improve this article if you can. (September 2010) | small = | smallimage = | smallimageright = | smalltext = }}
- [math]\displaystyle{ \begin{align} \phi(\mathbf{W}) = \frac{|\mathbf{S}_{B}^{\ast}|}{|\mathbf{S}_{W}^{\ast}|} = \frac{|\mathbf{W}^{T}\mathbf{S}_{B}\mathbf{W}|}{|\mathbf{W}^{T}\mathbf{S}_{W}\mathbf{W}|} \end{align} }[/math]
The solution for this question is that the columns of the transformation matrix [math]\displaystyle{ \mathbf{W} }[/math] are exactly the eigenvectors that correspond to largest [math]\displaystyle{ k-1 }[/math] eigenvalues with respect to
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. Please improve this article if you can. (What if we encounter complex eigenvalues? Then concept of being large does not dense. What is the solution in that case?) | small = | smallimage = | smallimageright = | smalltext = }}
- [math]\displaystyle{ \begin{align} \mathbf{S}_{W}^{-1}\mathbf{S}_{B}\mathbf{w}_{i} = \lambda_{i}\mathbf{w}_{i} \end{align} }[/math]
Also, note that we can use
- [math]\displaystyle{ \begin{align} \sum_{i=1}^{k}n_{i}\|(\mathbf{W}^{T}\mathbf{\mu}_{i}-\mathbf{W}^{T}\mathbf{\mu})^{T}\|^{2} \end{align} }[/math]
as our measure.
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: Please confirm that the identity below is true. Isn't the Euclidean matrix norm the square root of the larget eigenvalue of [math]\displaystyle{ X^*X }[/math]? Yes this is true the Euclidean matrix norm is the largest singular value of X. The square root of the trace of [math]\displaystyle{ X^*X }[/math] is the Frobenius Norm. See cleanup below for more info. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }}
Recall that
- [math]\displaystyle{ \begin{align} \|\mathbf{X}\|^2_{2} = Tr(\mathbf{X}^{T}\mathbf{X}) \end{align} }[/math]
Thus we obtain that {{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: Please confirm that the identity below is true.isn't it the Euclidean distance? why do yo continue with Frobenius distance?. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }}
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: The trace of [math]\displaystyle{ X^{T}X }[/math] is actually equal to the frobenius norm which is also the "entry-wise" 2 norm. The above norm is not the regular 2 norm which is the largest singular value of X. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }}
- [math]\displaystyle{ \begin{align} & \sum_{i=1}^{k}n_{i}\|(\mathbf{W}^{T}\mathbf{\mu}_{i}-\mathbf{W}^{T}\mathbf{\mu})^{T}\|^{2} \\ & = \sum_{i=1}^{k}n_{i}Tr[(\mathbf{W}^{T}\mathbf{\mu}_{i}-\mathbf{W}^{T}\mathbf{\mu})(\mathbf{W}^{T}\mathbf{\mu}_{i}-\mathbf{W}^{T}\mathbf{\mu})^{T}] \\ & = Tr[\sum_{i=1}^{k}n_{i}(\mathbf{W}^{T}\mathbf{\mu}_{i}-\mathbf{W}^{T}\mathbf{\mu})(\mathbf{W}^{T}\mathbf{\mu}_{i}-\mathbf{W}^{T}\mathbf{\mu})^{T}] \\ & = Tr[\sum_{i=1}^{k}n_{i}\mathbf{W}^{T}(\mathbf{\mu}_{i}-\mathbf{\mu})(\mathbf{\mu}_{i}-\mathbf{\mu})^{T}\mathbf{W}] \\ & = Tr[\mathbf{W}^{T}\sum_{i=1}^{k}n_{i}(\mathbf{\mu}_{i}-\mathbf{\mu})(\mathbf{\mu}_{i}-\mathbf{\mu})^{T}\mathbf{W}] \\ & = Tr[\mathbf{W}^{T}\mathbf{S}_{B}\mathbf{W}] \end{align} }[/math]
Similarly, we can get [math]\displaystyle{ Tr[\mathbf{W}^{T}\mathbf{S}_{W}\mathbf{W}] }[/math]. Thus we have following classic criterion function that Fisher used
- [math]\displaystyle{ \begin{align} \phi(\mathbf{W}) = \frac{Tr[\mathbf{W}^{T}\mathbf{S}_{B}\mathbf{W}]}{Tr[\mathbf{W}^{T}\mathbf{S}_{W}\mathbf{W}]} \end{align} }[/math]
Similar to the two class case problem, we have:
max [math]\displaystyle{ Tr[\mathbf{W}^{T}\mathbf{S}_{B}\mathbf{W}] }[/math] subject to [math]\displaystyle{ Tr[\mathbf{W}^{T}\mathbf{S}_{W}\mathbf{W}]=1 }[/math]
To solve this optimization problem a Lagrange multiplier [math]\displaystyle{ \Lambda }[/math], which actually is a [math]\displaystyle{ d \times d }[/math] diagonal matrix, is introduced:
- [math]\displaystyle{ \begin{align} L(\mathbf{W},\Lambda) = Tr[\mathbf{W}^{T}\mathbf{S}_{B}\mathbf{W}] - \Lambda\left\{ Tr[\mathbf{W}^{T}\mathbf{S}_{W}\mathbf{W}] - 1 \right\} \end{align} }[/math]
Differentiating with respect to [math]\displaystyle{ \mathbf{W} }[/math] we obtain:
- [math]\displaystyle{ \begin{align} \frac{\partial L}{\partial \mathbf{W}} = (\mathbf{S}_{B} + \mathbf{S}_{B}^{T})\mathbf{W} - \Lambda (\mathbf{S}_{W} + \mathbf{S}_{W}^{T})\mathbf{W} \end{align} }[/math]
Note that the [math]\displaystyle{ \mathbf{S}_{B} }[/math] and [math]\displaystyle{ \mathbf{S}_{W} }[/math] are both symmetric matrices, thus set the first derivative to zero, we obtain:
- [math]\displaystyle{ \begin{align} \mathbf{S}_{B}\mathbf{W} - \Lambda\mathbf{S}_{W}\mathbf{W}=0 \end{align} }[/math]
Thus,
- [math]\displaystyle{ \begin{align} \mathbf{S}_{B}\mathbf{W} = \Lambda\mathbf{S}_{W}\mathbf{W} \end{align} }[/math]
where
- [math]\displaystyle{ \mathbf{\Lambda} = \begin{pmatrix} \lambda_{1} & & 0\\ &\ddots&\\ 0 & &\lambda_{d} \end{pmatrix} }[/math]
and [math]\displaystyle{ \mathbf{W} = [\mathbf{w}_{1}, \mathbf{w}_{2},..., \mathbf{w}_{k-1}] }[/math].
As a matter of fact, [math]\displaystyle{ \mathbf{\Lambda} }[/math] must have [math]\displaystyle{ \mathbf{k-1} }[/math] nonzero eigenvalues, because [math]\displaystyle{ rank({S}_{W}^{-1}\mathbf{S}_{B})=k-1 }[/math].
Therefore, the solution for this question is as same as the previous case. The columns of the transformation matrix [math]\displaystyle{ \mathbf{W} }[/math] are exactly the eigenvectors that correspond to largest [math]\displaystyle{ k-1 }[/math] eigenvalues with respect to
- [math]\displaystyle{ \begin{align} \mathbf{S}_{W}^{-1}\mathbf{S}_{B}\mathbf{w}_{i} = \lambda_{i}\mathbf{w}_{i} \end{align} }[/math]
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: Adding more general comments about the advantages and flaws of FDA would be effective here.. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }}
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: Would you please show how could we reconstruct our original data from the data that its dimentionality is reduced by FDA.. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }} {{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: When you reduce the dimensionality of data in most general form you lose some features of the data and you cannot reconstruct the data from redacted space unless the data have special features that help you in reconstruction like sparsity. In FDA it seems that we cannot reconstruct data in general form using reducted version of data. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }}
Generalization of Fisher's Linear Discriminant Analysis
Fisher's Linear Discriminant Analysis (Fisher, 1936) is very popular among users of discriminant analysis. Some of the reasons for this are its simplicity and lack of necessity for strict assumptions. However, it has optimality properties only if the underlying distributions of the groups are multivariate normal. It is also easy to verify that the discriminant rule obtained can be very harmed by only a small number of outlying observations. Outliers are very hard to detect in multivariate data sets and even when they are detected simply discarding them is not the most efficient way of handling the situation. Therefore, there is a need for robust procedures that can accommodate the outliers and are not strongly affected by them. Then, a generalization of Fisher's linear discriminant algorithm [[10]]is developed to lead easily to a very robust procedure.
Also notice that LDA can be seen as a dimensionality reduction technique. In general k-class problems, we have k means which lie on a linear subspace with dimension k-1. Given a data point, we are looking for the closest class mean to this point. In LDA, we project the data point to the linear subspace and calculate distances within that subspace. If the dimensionality of the data, d, is much larger than the number of classes, k, then we have a considerable drop in dimension.
Linear and Logistic Regression - October 12, 2010
Linear Regression
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: It would be nice if someone explains the relation between linear regression and classification. Why did we switch to linear regression which seems to be a different topic. Please improve this article if you can. (September 2010) | small = | smallimage = | smallimageright = | smalltext = }}
We will start by considering a very simple regression model, the linear regression model.
According to Bayes Classification,
[math]\displaystyle{ P( Y=k | X=x )= \frac{f_{k}(x)\pi_{k}}{\Sigma_{k}f_{k}(x)\pi_{k}} }[/math]
For the purpose of classification, the linear regression model assumes that the regression function [math]\displaystyle{ \,E(Y|X) }[/math] is linear in the inputs [math]\displaystyle{ \,\mathbf{x}_{1}, ..., \mathbf{x}_{p} }[/math].
The simple linear regression model has the general form:
- [math]\displaystyle{ \begin{align} y_i = \beta^{T}\mathbf{x}_{i}+\beta_{0} \end{align} }[/math]
and we can denote it as
- [math]\displaystyle{ \begin{align} \mathbf{y} = \beta^{T}\mathbf{X} \end{align} }[/math]
where [math]\displaystyle{ \,\beta^{T} = ( \beta_1,..., \beta_{d},\beta_0) }[/math] is a [math]\displaystyle{ 1 \times (d+1) }[/math] vector and [math]\displaystyle{ \mathbf{X}= \begin{pmatrix} \mathbf{x}_{1}, \dots,\mathbf{x}_{n}\\ 1, \dots, 1 \end{pmatrix} }[/math] is a [math]\displaystyle{ (d+1) \times n }[/math] Matrix,here [math]\displaystyle{ \mathbf{x}_{i} }[/math] is a [math]\displaystyle{ d \times 1 }[/math] vector
Given input data [math]\displaystyle{ \,\mathbf{x}_{1}, ..., \mathbf{x}_{n} }[/math] and [math]\displaystyle{ \,y_{1}, ..., y_{n} }[/math] our goal is to find [math]\displaystyle{ \,\beta^{T} }[/math] such that the linear model fits the data while minimizing sum of squared errors using the Least Squares method.
Note that vectors [math]\displaystyle{ \mathbf{x}_{i} }[/math] could be numerical inputs, transformations of the original data, i.e. [math]\displaystyle{ \log \mathbf{x}_{i} }[/math] or [math]\displaystyle{ \sin \mathbf{x}_{i} }[/math], or basis expansions, i.e. [math]\displaystyle{ \mathbf{x}_{i}^{2} }[/math] or [math]\displaystyle{ \mathbf{x}_{i}\times \mathbf{x}_{j} }[/math].
We then try to minimize the residual sum-of-squares
- [math]\displaystyle{ \begin{align} \mathrm{RSS}(\beta)=(\mathbf{y}-\beta^{T}\mathbf{X})^{T}(\mathbf{y}-\beta^{T}\mathbf{X}) \end{align} }[/math]
This is a quadratic function in the [math]\displaystyle{ \,d+1 }[/math] parameters. Differentiating with respect to [math]\displaystyle{ \,\beta }[/math] we obtain
- [math]\displaystyle{ \begin{align} \frac{\partial \mathrm{RSS}}{\partial \beta} = -2\mathbf{X}(\mathbf{y}-\beta^{T}\mathbf{X})^{T} \end{align} }[/math]
- [math]\displaystyle{ \begin{align} \frac{\partial^{2}\mathrm{RSS}}{\partial \beta \partial \beta^{T}}=2\mathbf{X}^{T}\mathbf{X} \end{align} }[/math]
Set the first derivative to zero
- [math]\displaystyle{ \begin{align} \mathbf{X}(\mathbf{y}^{T}-\mathbf{X}^{T}\beta)=0 \end{align} }[/math]
we obtain the solution
- [math]\displaystyle{ \begin{align} \hat \beta = (\mathbf{X}\mathbf{X}^{T})^{-1}\mathbf{X}\mathbf{y}^{T} \end{align} }[/math]
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: we use :[math]\displaystyle{ \begin{align} \mathbf{y} = \beta^{T}\mathbf{X} \end{align} }[/math] in this course, but
- [math]\displaystyle{ \begin{align} \mathbf{y} = \mathbf{X}\beta \end{align} }[/math] were used by the notes last year and then it has the result below. I am confused these results seem to hold if we use [math]\displaystyle{ \beta^{T}X }[/math] rather than [math]\displaystyle{ X\Beta }[/math]. If we use [math]\displaystyle{ X\Beta }[/math] then X has to be a n x (d+1) matrix and [math]\displaystyle{ \Beta }[/math] is a (d+1) x 1 vector. Has this cleanup been fixed or does it still stand?. Please improve this article if you can. (12 Oct 2010)
| small = | smallimage = | smallimageright = | smalltext = }} {{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: It depends on the definition of :[math]\displaystyle{ \begin{align} \beta^{T}\end{align} }[/math] and/or [math]\displaystyle{ \begin{align}\beta\end{align} }[/math] as the second is nothing but the first, added to it :[math]\displaystyle{ \begin{align}\beta0\end{align} }[/math], then :[math]\displaystyle{ \begin{align}\beta\end{align} }[/math] = [[math]\displaystyle{ \begin{align} \beta0\end{align} }[/math] [math]\displaystyle{ \begin{align}\beta^{T}\end{align} }[/math]] I think this is close to the right explanation, please make sure of it and post the right thing. Please improve this article if you can. (12 Oct 2010) | small = | smallimage = | smallimageright = | smalltext = }}
Thus the fitted values at the inputs are
- [math]\displaystyle{ \begin{align} \mathbf{\hat y} = \hat\beta^T\mathbf{X} = \mathbf{y}\mathbf{X}^{T}(\mathbf{X}^{T}\mathbf{X})^{-1}\mathbf{X} \end{align} }[/math]
where [math]\displaystyle{ \mathbf{H} = \mathbf{X}^{T}(\mathbf{X}^{T}\mathbf{X})^{-1}\mathbf{X} }[/math] is called the hat matrix.
- Note For classification purposes, this is not a correct model. Recall the following application of Bayes classifier:
[math]\displaystyle{ r(x)= P( Y=k | X=x )= \frac{f_{k}(x)\pi_{k}}{\Sigma_{k}f_{k}(x)\pi_{k}} }[/math]
It is clear that to make sense mathematically, [math]\displaystyle{ \displaystyle r(x) }[/math] must be a value between 0 and 1. If this is estimated with the
regression function [math]\displaystyle{ \displaystyle r(x)=E(Y|X=x) }[/math] and [math]\displaystyle{ \mathbf{\hat\beta} }[/math] is learned as above, then there is nothing that would restrict [math]\displaystyle{ \displaystyle r(x) }[/math] to taking values between 0 and 1. This is more direct approach to classification since it do not need to estimate [math]\displaystyle{ \ f_k(x) }[/math] and [math]\displaystyle{ \ \pi_k }[/math].
[math]\displaystyle{ \ 1 \times P(Y=1|X=x)+0 \times P(Y=0|X=x)=E(Y|X) }[/math]
This model does not classify Y between 0 and 1, so it is not good and sometimes it can lead to a decent classifier. [math]\displaystyle{ \ y_i=\frac{1}{n_1} }[/math] [math]\displaystyle{ \ \frac{-1}{n_2} }[/math]
Logistic Regression
The logistic regression model arises from the desire to model the posterior probabilities of the [math]\displaystyle{ \displaystyle K }[/math] classes via linear functions in [math]\displaystyle{ \displaystyle x }[/math], while at the same time ensuring that they sum to one and remain in [0,1].Logistic regression models are usually fit by maximum likelihood, using the conditional likelihood ,using [math]\displaystyle{ \displaystyle Pr(Y|X) }[/math]. Since [math]\displaystyle{ \displaystyle Pr(Y|X) }[/math] completely specifies the conditional distribution, the multinomial distribution is appropriate. This model is widely used in biostatistical applications for two classes. For instance: people survive or die, have a disease or not, have a risk factor or not.
logistic function
A logistic function or logistic curve is the most common sigmoid curve.
1. [math]\displaystyle{ y = \frac{1}{1+e^{-x}} }[/math]
2. [math]\displaystyle{ \frac{dy}{dx} = y(1-y)=\frac{e^{x}}{(1+e^{x})^{2}} }[/math]
3. [math]\displaystyle{ y(0) = \frac{1}{2} }[/math]
4. [math]\displaystyle{ \int y dx = ln(1 + e^{x}) }[/math]
5. [math]\displaystyle{ y(x) = \frac{1}{2} + \frac{1}{4}x - \frac{1}{48}x^{3} + \frac{1}{48}x^{5} \cdots }[/math]
The logistic curve shows early exponential growth for negative t, which slows to linear growth of slope 1/4 near t = 0, then approaches y = 1 with an exponentially decaying gap.
Intuition behind Logistic Regression
Recall that, for classification purposes, the linear regression model presented in the above section is not correct because it does not force [math]\displaystyle{ \,r(x) }[/math] to be between 0 and 1 and sum to 1. Consider the following log odds model (for two classes):
- [math]\displaystyle{ \log\left(\frac{P(Y=1|X=x)}{P(Y=0|X=x)}\right)=\beta^Tx }[/math]
Calculating [math]\displaystyle{ \,P(Y=1|X=x) }[/math] leads us to the logistic regression model, which as opposed to the linear regression model, allows the modelling of the posterior probabilities of the classes through linear methods and at the same time ensures that they sum to one and are between 0 and 1. It is a type of Generalized Linear Model (GLM).
The Logistic Regression Model
The logistic regression model for the two class case is defined as
Class 1 {{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: It could be useful to have sources for these graphs. We don't know if they are copyrighted. Please improve this article if you can. (October 13 2010) | small = | smallimage = | smallimageright = | smalltext = }}

- [math]\displaystyle{ P(Y=1 | X=x) =\frac{\exp(\underline{\beta}^T \underline{x})}{1+\exp(\underline{\beta}^T \underline{x})}=P(x;\underline{\beta}) }[/math]
Then we have that
Class 0 {{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: It could be useful to have sources for these graphs. We don't know if they are copyrighted. Please improve this article if you can. (October 13 2010) | small = | smallimage = | smallimageright = | smalltext = }}

- [math]\displaystyle{ P(Y=0 | X=x) = 1-P(Y=1 | X=x)=1-\frac{\exp(\underline{\beta}^T \underline{x})}{1+\exp(\underline{\beta}^T \underline{x})}=\frac{1}{1+\exp(\underline{\beta}^T \underline{x})} }[/math]
Fitting a Logistic Regression
Logistic regression tries to fit a distribution. The common practice in statistics is to fit density function, posterior density of each class(Pr(Y|X), to data using maximum likelihood. The maximum likelihood of [math]\displaystyle{ \underline\beta }[/math] maximizes the probability of obtaining the data [math]\displaystyle{ \displaystyle{x_{1},...,x_{n}} }[/math] from the known distribution. Combining [math]\displaystyle{ \displaystyle P(Y=1 | X=x) }[/math] and [math]\displaystyle{ \displaystyle P(Y=0 | X=x) }[/math] as follows, we can consider the two classes at the same time:
- [math]\displaystyle{ p(\underline{x_{i}};\underline{\beta}) = \left(\frac{\exp(\underline{\beta}^T \underline{x_i})}{1+\exp(\underline{\beta}^T \underline{x_i})}\right)^{y_i} \left(\frac{1}{1+\exp(\underline{\beta}^T \underline{x_i})}\right)^{1-y_i} }[/math]
Assuming the data [math]\displaystyle{ \displaystyle {x_{1},...,x_{n}} }[/math] is drawn independently, the likelihood function is
- [math]\displaystyle{ \begin{align} \mathcal{L}(\theta)&=p({x_{1},...,x_{n}};\theta)\\ &=\displaystyle p(x_{1};\theta) p(x_{2};\theta)... p(x_{n};\theta) \quad \mbox{(by independence)}\\ &= \prod_{i=1}^n p(x_{i};\theta) \end{align} }[/math]
Since it is more convenient to work with the log-likelihood function, take the log of both sides, we get
- [math]\displaystyle{ \displaystyle l(\theta)=\displaystyle \sum_{i=1}^n \log p(x_{i};\theta) }[/math]
So,
- [math]\displaystyle{ \begin{align} l(\underline\beta)&=\displaystyle\sum_{i=1}^n y_{i}\log\left(\frac{\exp(\underline{\beta}^T \underline{x_i})}{1+\exp(\underline{\beta}^T \underline{x_i})}\right)+(1-y_{i})\log\left(\frac{1}{1+\exp(\underline{\beta}^T\underline{x_i})}\right)\\ &= \displaystyle\sum_{i=1}^n y_{i}(\underline{\beta}^T\underline{x_i}-\log(1+\exp(\underline{\beta}^T\underline{x_i}))+(1-y_{i})(-\log(1+\exp(\underline{\beta}^T\underline{x_i}))\\ &= \displaystyle\sum_{i=1}^n y_{i}\underline{\beta}^T\underline{x_i}-y_{i} \log(1+\exp(\underline{\beta}^T\underline{x_i}))- \log(1+\exp(\underline{\beta}^T\underline{x_i}))+y_{i} \log(1+\exp(\underline{\beta}^T\underline{x_i}))\\ &=\displaystyle\sum_{i=1}^n y_{i}\underline{\beta}^T\underline{x_i}- \log(1+\exp(\underline{\beta}^T\underline{x_i}))\\ \end{align} }[/math]
To maximize the log-likelihood, set its derivative to 0.
- [math]\displaystyle{ \begin{align} \frac{\partial l}{\partial \underline{\beta}} &= \sum_{i=1}^n \left[{y_i} \underline{x}_i- \frac{\exp(\underline{\beta}^T \underline{x_i})}{1+\exp(\underline{\beta}^T \underline{x}_i)}\underline{x}_i\right]\\ &=\sum_{i=1}^n \left[{y_i} \underline{x}_i - p(\underline{x}_i;\underline{\beta})\underline{x}_i\right] \end{align} }[/math]
There are n+1 nonlinear equations in [math]\displaystyle{ / \beta }[/math]. The first column is vector 1, then [math]\displaystyle{ \ \sum_{i=1}^n {y_i} =\sum_{i=1}^n p(\underline{x}_i;\underline{\beta}) }[/math] i.e. the expected number of class ones matches the observed number.
To solve this equation, the Newton-Raphson algorithm is used which requires the second derivative in addition to the first derivative. This is demonstrated in the next section.
Extension
- When we are dealing with a problem with more than two classes, we need to generalize our logistic regression to a Multinomial Logit model.
- Limitations of Logistic Regression:
- 1. We know that there is no assumptions are made about the distributions of the features of the data (i.e. the explanatory variables). However, the features should not be highly correlated with one another because this could cause problems with estimation.
- 2. Large number of data points (i.e.the sample sizes) are required for logistic regression to provide sufficient numbers in both classes. The more number of features/dimensions of the data, the larger the sample size required.
Logistic Regression Cont. - October 14, 2010
Logistic Regression Model
Recall that in the last lecture, we learned the logistic regression model.
- [math]\displaystyle{ P(Y=1 | X=x)=P(\underline{x}_i;\underline{\beta})=\frac{exp(\underline{\beta}^T \underline{x})}{1+exp(\underline{\beta}^T \underline{x})} }[/math]
- [math]\displaystyle{ P(Y=0 | X=x)=1-P(\underline{x}_i;\underline{\beta})=\frac{1}{1+exp(\underline{\beta}^T \underline{x})} }[/math]
Estimating Parameters [math]\displaystyle{ \underline{\beta} }[/math]
Criteria: find a [math]\displaystyle{ \underline{\beta} }[/math] that maximizes the conditional likelihood of Y given X using the training data.
From above, we have the first derivative of the log-likelihood:
[math]\displaystyle{ \frac{\partial l}{\partial \underline{\beta}} = \sum_{i=1}^n \left[{y_i} \underline{x}_i- \frac{exp(\underline{\beta}^T \underline{x_i})}{1+exp(\underline{\beta}^T \underline{x}_i)}\underline{x}_i\right] }[/math] [math]\displaystyle{ =\sum_{i=1}^n \left[{y_i} \underline{x}_i - P(\underline{x}_i;\underline{\beta})\underline{x}_i\right] }[/math]
Newton-Raphson Algorithm:
If we want to find [math]\displaystyle{ \ x^* }[/math] such that [math]\displaystyle{ \ f(x^*)=0 }[/math]
We first pick a starting point [math]\displaystyle{ x^* = x^{old} }[/math] and and we solve:
[math]\displaystyle{ \ x^{*} \leftarrow x^{old}-\frac {f(x^{old})}{\partial f(x^{old})} }[/math]
[math]\displaystyle{ \ x^{old} \leftarrow x^{*} }[/math]
This is repeated till convergence
If we want to maximize or minimize [math]\displaystyle{ \ f(x) }[/math], then solve for [math]\displaystyle{ \ \partial f(x)=0 }[/math]
[math]\displaystyle{ \ X^{new} \leftarrow x^{old}-\frac {\partial f(x^{old})}{\partial^2 f(x^{old})} }[/math]
In vector notation the above can be written as
[math]\displaystyle{
X^{new} \leftarrow X^{old} - H^{-1}\Delta
}[/math]
H is the Hessian matrix or second derivative matrix and [math]\displaystyle{ \Delta }[/math] is the gradient both evaluated at [math]\displaystyle{ X^{old} }[/math]
note: If the Hessian is not invertible the generalized inverse or pseudo inverse can be used
As shown above ,the Newton-Raphson algorithm requires the second-derivative or Hessian.
[math]\displaystyle{ \frac{\partial^{2} l}{\partial \underline{\beta} \partial \underline{\beta}^T }= \sum_{i=1}^n - \underline{x_i} \frac{(exp(\underline{\beta}^T\underline{x}_i) \underline{x}_i^T)(1+exp(\underline{\beta}^T \underline{x}_i))-\underline{x}_i exp(\underline{\beta}^T\underline{x}_i)exp(\underline{\beta}^T\underline{x}_i)}{(1+exp(\underline{\beta}^T \underline{x}))^2} }[/math]
(note: [math]\displaystyle{ \frac{\partial\underline{\beta}^T\underline{x}_i}{\partial \underline{\beta}^T}=\underline{x}_i^T }[/math] you can check it here, it's a very useful website including a Matrix Reference Manual that you can find information about linear algebra and the properties of real and complex matrices.)
- [math]\displaystyle{ =\sum_{i=1}^n - \underline{x}_i \frac{(-\underline{x}_i exp(\underline{\beta}^T\underline{x}_i) \underline{x}_i^T)}{(1+exp(\underline{\beta}^T \underline{x}))(1+exp(\underline{\beta}^T \underline{x}))} }[/math] (by cancellation)
- [math]\displaystyle{ =\sum_{i=1}^n - \underline{x}_i \underline{x}_i^T P(\underline{x}_i;\underline{\beta}))[1-P(\underline{x}_i;\underline{\beta})]) }[/math](since [math]\displaystyle{ P(\underline{x}_i;\underline{\beta})=\frac{exp(\underline{\beta}^T \underline{x})}{1+exp(\underline{\beta}^T \underline{x})} }[/math] and [math]\displaystyle{ 1-P(\underline{x}_i;\underline{\beta})=\frac{1}{1+exp(\underline{\beta}^T \underline{x})} }[/math])
The same second derivative can be achieved if we reduce the occurrences of beta to 1 by the identity[math]\displaystyle{ \frac{a}{1+a}=1-\frac{1}{1+a} }[/math]
And solving [math]\displaystyle{ \frac{\partial}{\partial \underline{\beta}^T}\sum_{i=1}^n \left[{y_i} \underline{x}_i-\left[1-\frac{1}{1+exp(\underline{\beta}^T \underline{x}_i)}\right]\underline{x}_i\right] }[/math]
Starting with [math]\displaystyle{ \,\underline{\beta}^{old} }[/math], the Newton-Raphson update is
[math]\displaystyle{ \,\underline{\beta}^{new}\leftarrow \,\underline{\beta}^{old}- (\frac{\partial ^2 l}{\partial \underline{\beta}\partial \underline{\beta}^T})^{-1}(\frac{\partial l}{\partial \underline{\beta}}) }[/math] where the derivatives are evaluated at [math]\displaystyle{ \,\underline{\beta}^{old} }[/math]
The iteration will terminate when [math]\displaystyle{ \underline{\beta}^{new} }[/math] is very close to [math]\displaystyle{ \underline{\beta}^{old} }[/math].
The iteration can be described in matrix form.
- Let [math]\displaystyle{ \,\underline{Y} }[/math] be the column vector of [math]\displaystyle{ \,y_i }[/math]. ([math]\displaystyle{ n\times1 }[/math])
- Let [math]\displaystyle{ \,X }[/math] be the [math]\displaystyle{ {d}\times{n} }[/math] input matrix.
- Let [math]\displaystyle{ \,\underline{P} }[/math] be the [math]\displaystyle{ {n}\times{1} }[/math] vector with [math]\displaystyle{ i }[/math]th element [math]\displaystyle{ P(\underline{x}_i;\underline{\beta}^{old}) }[/math].
- Let [math]\displaystyle{ \,W }[/math] be an [math]\displaystyle{ {n}\times{n} }[/math] diagonal matrix with [math]\displaystyle{ i }[/math]th element [math]\displaystyle{ P(\underline{x}_i;\underline{\beta}^{old})[1-P(\underline{x}_i;\underline{\beta}^{old})] }[/math]
then
[math]\displaystyle{ \frac{\partial l}{\partial \underline{\beta}} = X(\underline{Y}-\underline{P}) }[/math]
[math]\displaystyle{ \frac{\partial ^2 l}{\partial \underline{\beta}\partial \underline{\beta}^T} = -XWX^T }[/math]
The Newton-Raphson step is
[math]\displaystyle{ \underline{\beta}^{new} \leftarrow \underline{\beta}^{old}+(XWX^T)^{-1}X(\underline{Y}-\underline{P}) }[/math]
This equation is sufficient for computation of the logistic regression model. However, we can simplify further to uncover an interesting feature of this equation.
[math]\displaystyle{ \begin{align} \underline{\beta}^{new} &= (XWX^T)^{-1}(XWX^T)\underline{\beta}^{old}+(XWX^T)^{-1}XWW^{-1}(\underline{Y}-\underline{P})\\ &=(XWX^T)^{-1}XW[X^T\underline{\beta}^{old}+W^{-1}(\underline{Y}-\underline{P})]\\ &=(XWX^T)^{-1}XWZ \end{align} }[/math]
where [math]\displaystyle{ Z=X\underline{\beta}^{old}+W^{-1}(\underline{Y}-\underline{P}) }[/math]
This is a adjusted response and it is solved repeatedly when [math]\displaystyle{ \ p }[/math], [math]\displaystyle{ \ W }[/math], and [math]\displaystyle{ \ z }[/math] changes. This algorithm is called iteratively reweighted least squares because it solves the weighted least squares problem repeatedly.
Recall that linear regression by least square finds the following minimum: [math]\displaystyle{ \min_{\underline{\beta}}(\underline{y}-\underline{\beta}^T X)^T(\underline{y}-\underline{\beta}^TX) }[/math]
we have [math]\displaystyle{ \underline\hat{\beta}=(XX^T)^{-1}X\underline{y} }[/math]
Similarly, we can say that [math]\displaystyle{ \underline{\beta}^{new} }[/math] is the solution of a weighted least square problem:
[math]\displaystyle{ \underline{\beta}^{new} \leftarrow arg \min_{\underline{\beta}}(Z-X\underline{\beta}^T)W(Z-X\underline{\beta}) }[/math]
Pseudo Code
- [math]\displaystyle{ \underline{\beta} \leftarrow 0 }[/math]
- Set [math]\displaystyle{ \,\underline{Y} }[/math], the label associated with each observation [math]\displaystyle{ \,i=1...n }[/math].
- Compute [math]\displaystyle{ \,\underline{P} }[/math] according to the equation [math]\displaystyle{ P(\underline{x}_i;\underline{\beta})=\frac{exp(\underline{\beta}^T \underline{x})}{1+exp(\underline{\beta}^T \underline{x})} }[/math] for all [math]\displaystyle{ \,i=1...n }[/math].
- Compute the diagonal matrix [math]\displaystyle{ \,W }[/math] by setting [math]\displaystyle{ \,w_i,i }[/math] to [math]\displaystyle{ P(\underline{x}_i;\underline{\beta}))[1-P(\underline{x}_i;\underline{\beta})] }[/math] for all [math]\displaystyle{ \,i=1...n }[/math].
- [math]\displaystyle{ Z \leftarrow X^T\underline{\beta}+W^{-1}(\underline{Y}-\underline{P}) }[/math].
- [math]\displaystyle{ \underline{\beta} \leftarrow (XWX^T)^{-1}XWZ }[/math].
- If the new [math]\displaystyle{ \underline{\beta} }[/math] value is sufficiently close to the old value, stop; otherwise go back to step 3.
Comparison with Linear Regression
- Similarities
- They are both to attempt to estimate [math]\displaystyle{ \,P(Y=k|X=x) }[/math] (For logistic regression, we just mentioned about the case that [math]\displaystyle{ \,k=0 }[/math] or [math]\displaystyle{ \,k=1 }[/math] now).
- They are both have linear boundaries.
- note:For linear regression, we assume the model is linear. The boundary is [math]\displaystyle{ P(Y=k|X=x)=\underline{\beta}^T\underline{x}_i+\underline{\beta}_0=0.5 }[/math] (linear)
- For logistic regression, the boundary is [math]\displaystyle{ P(Y=k|X=x)=\frac{exp(\underline{\beta}^T \underline{x})}{1+exp(\underline{\beta}^T \underline{x})}=0.5 \Rightarrow exp(\underline{\beta}^T \underline{x})=1\Rightarrow \underline{\beta}^T \underline{x}=0 }[/math] (linear)
- Differences
- Linear regression: [math]\displaystyle{ \,P(Y=k|X=x) }[/math] is linear function of [math]\displaystyle{ \,x }[/math], [math]\displaystyle{ \,P(Y=k|X=x) }[/math] is not guaranteed to fall between 0 and 1 and to sum up to 1. There exists a closed form solution for least squares.
- Logistic regression: [math]\displaystyle{ \,P(Y=k|X=x) }[/math] is a nonlinear function of [math]\displaystyle{ \,x }[/math], and it is guaranteed to range from 0 to 1 and to sum up to 1. No closed form solution exists
Comparison with LDA
- The linear logistic model only consider the conditional distribution [math]\displaystyle{ \,P(Y=k|X=x) }[/math]. No assumption is made about [math]\displaystyle{ \,P(X=x) }[/math].
- The LDA model specifies the joint distribution of [math]\displaystyle{ \,X }[/math] and [math]\displaystyle{ \,Y }[/math].
- Logistic regression maximizes the conditional likelihood of [math]\displaystyle{ \,Y }[/math] given [math]\displaystyle{ \,X }[/math]: [math]\displaystyle{ \,P(Y=k|X=x) }[/math]
- LDA maximizes the joint likelihood of [math]\displaystyle{ \,Y }[/math] and [math]\displaystyle{ \,X }[/math]: [math]\displaystyle{ \,P(Y=k,X=x) }[/math].
- If [math]\displaystyle{ \,\underline{x} }[/math] is d-dimensional,the number of adjustable parameter in logistic regression is [math]\displaystyle{ \,d }[/math]. The number of parameters grows linearly w.r.t dimension.
- If [math]\displaystyle{ \,\underline{x} }[/math] is d-dimensional,the number of adjustable parameter in LDA is [math]\displaystyle{ \,(2d)+d(d+1)/2+2=(d^2+5d+4)/2 }[/math]. The number of parameters grows quadratically w.r.t dimension.
- LDA estimate parameters more efficiently by using more information about data and samples without class labels can be also used in LDA.
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: Could somebody please validate the following points. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }}
- As logistic regression relies on fewer assumptions, it seems to be more robust.
- In practice, Logistic regression and LDA often give the similar results.
By example
Now we compare LDA and Logistic regression by an example. Again, we use them on the 2_3 data.
>>load 2_3; >>[U, sample] = princomp(X'); >>sample = sample(:,1:2); >>plot (sample(1:200,1), sample(1:200,2), '.'); >>hold on; >>plot (sample(201:400,1), sample(201:400,2), 'r.');
- First, we do PCA on the data and plot the data points that represent 2 or 3 in different colors. See the previous example for more details.
>>group = ones(400,1); >>group(201:400) = 2;
- Group the data points.
>>[B,dev,stats] = mnrfit(sample,group); >>x=[ones(1,400); sample'];
- Now we use mnrfit to use logistic regression to classfy the data. This function can return B which is a [math]\displaystyle{ (d+1)\times{(k–1)} }[/math] matrix of estimates, where each column corresponds to the estimated intercept term and predictor coefficients. In this case, B is a [math]\displaystyle{ 3\times{1} }[/math] matrix.
>> B B =0.1861 -5.5917 -3.0547
- This is our [math]\displaystyle{ \underline{\beta} }[/math]. So the posterior probabilities are:
- [math]\displaystyle{ P(Y=1 | X=x)=\frac{exp(0.1861-5.5917X_1-3.0547X_2)}{1+exp(0.1861-5.5917X_1-3.0547X_2)} }[/math].
- [math]\displaystyle{ P(Y=2 | X=x)=\frac{1}{1+exp(0.1861-5.5917X_1-3.0547X_2)} }[/math]
- The classification rule is:
- [math]\displaystyle{ \hat Y = 1 }[/math], if [math]\displaystyle{ \,0.1861-5.5917X_1-3.0547X_2\gt =0 }[/math]
- [math]\displaystyle{ \hat Y = 2 }[/math], if [math]\displaystyle{ \,0.1861-5.5917X_1-3.0547X_2\lt 0 }[/math]
>>f = sprintf('0 = %g+%g*x+%g*y', B(1), B(2), B(3));
>>ezplot(f,[min(sample(:,1)), max(sample(:,1)), min(sample(:,2)), max(sample(:,2))])
- Plot the decision boundary by logistic regression.
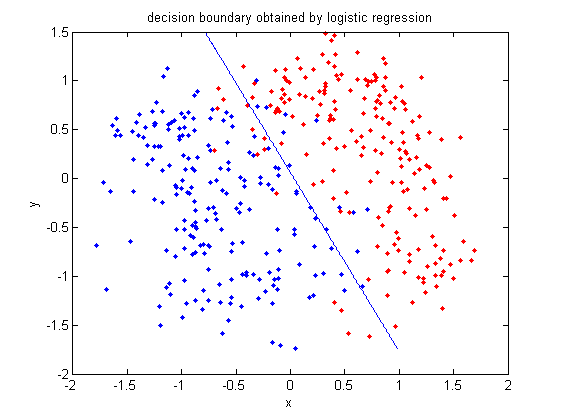
>>[class, error, POSTERIOR, logp, coeff] = classify(sample, sample, group, 'linear');
>>k = coeff(1,2).const;
>>l = coeff(1,2).linear;
>>f = sprintf('0 = %g+%g*x+%g*y', k, l(1), l(2));
>>h=ezplot(f, [min(sample(:,1)), max(sample(:,1)), min(sample(:,2)), max(sample(:,2))]);
- Plot the decision boundary by LDA. See the previous example for more information about LDA in matlab.
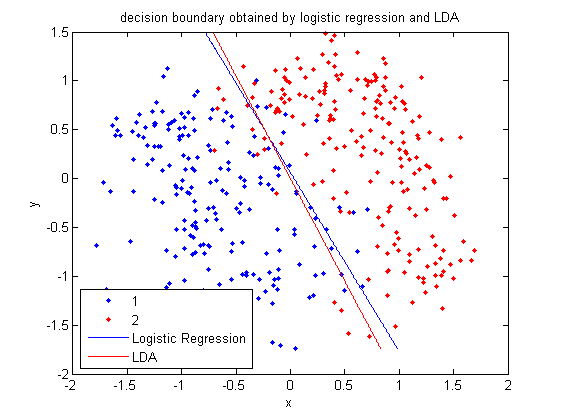
Lecture Summary
Traditionally logistic regression parameters are estimated using maximum likelihood. However , other optimization techniques may be used as well.
{{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: It would be nice if a link to a proof showing that logistic regression is convex and that Newtons will converge to a global. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }} {{
Template:namespace detect
| type = style | image = | imageright = | style = | textstyle = | text = This article may require cleanup to meet Wikicoursenote's quality standards. The specific problem is: I found a proof and I add the link in 'Supplements', please check if it is suitable or not. Please improve this article if you can. (October 2010) | small = | smallimage = | smallimageright = | smalltext = }}
Since there is no closed form solution for finding the zero of the first derivative of the log likelihood the Newton Raphson algorithm is used. Since the problem is convex Newtons is guaranteed to converge to a global optimum.
Logistic regression requires less parameters than LDA or QDA and is therefore more favorable for high dimensional data.
Supplements
A detailed proof that logistic regression is convex is available here. See '1 Binary LR' for the case we discussed in lecture.
- Pages with broken file links
- Articles needing cleanup from October 8 2010
- Articles with invalid date parameter in template
- All pages needing cleanup
- Articles needing cleanup from October 14 2010
- Articles needing cleanup from September 6 2010
- Articles needing cleanup from October 2010
- Articles needing cleanup from October2010
- Articles needing cleanup from September 2010
- Articles needing cleanup from What if we encounter complex eigenvalues? Then concept of being large does not dense. What is the solution in that case?
- Articles needing cleanup from 12 Oct 2010
- Articles needing cleanup from October 13 2010