deep Neural Nets as a Method for Quantitative Structure–Activity Relationships: Difference between revisions
No edit summary |
|||
| Line 95: | Line 95: | ||
[[File: table2.PNG | frame | center |Comparing RF with DNN trained using recommended parameter settings on 15 additional datasets]] | [[File: table2.PNG | frame | center |Comparing RF with DNN trained using recommended parameter settings on 15 additional datasets]] | ||
</center> | </center> | ||
Both RF and DNN can be efficiently speeded up using high performance computing technologies, but in a different way due to the inherent difference in their algorithms. | |||
== Discussion == | == Discussion == | ||
Revision as of 15:29, 23 November 2015
Introduction
This abstract is a summary of the paper "Deep Neural Nets as a Method for Quantitative Structure−Activity Relationships" by Ma J. et al. <ref> Ma J, Sheridan R. et al. [ http://pubs.acs.org/doi/pdf/10.1021/ci500747n.pdf "QSAR deep nets"] Journal of Chemical Information and Modeling. 2015,55, 263-274</ref>. The paper presents the application of machine learning methods, specifically Deep Neural Networks <ref> Hinton, G. E.; Osindero, S.; Teh, Y. W. A fast learning algorithm for deep belief nets. Neural Computation 2006, 18, 1527−1554</ref> and Random Forest models <ref> Breiman L. Random Forests, Machine Learning. 2001,45, 5-32</ref> in the field of pharmaceutical industry. To discover a drug, it is needed that the best combination of different chemical compounds with different molecular structure was selected in order to achieve the best biological activity. Currently the SAR (QSAR) models are routinely used for this purpose. Structure-Activity Relationship (SAR), or Quantified SAR, is an approach designed to find relationships between chemical structure and biological activity (or target property) of studied compounds. The SAR models are type of classification or regression models where the predictors consist of physio-chemical properties or theoretical molecular and the response variable could be a biological activity of the chemicals, such as concentration of a substance required to give a certain biological response. The basic idea behind these methods is that activity of molecules is reflected in their structure and same molecules have the same activity. So if we learn the activity of a set of molecules structures ( or combinations of molecules) then we can predict the activity of similar molecules. QSAR methods are particularly computer intensive or require the adjustment of many sensitive parameters to achieve good prediction.In this sense, the machine learning methods can be helpful and two of those methods: support vector machine (SVM) and random forest (RF) are commonly used <ref>Svetnik, V. et al.,[http://pubs.acs.org/doi/pdf/10.1021/ci034160g.pdf Random forest: a classification and regression tool for compound classification and QSAR modeling,J. Chem. Inf. Comput. Sci. 2003, 43, 1947−1958 </ref>. In this paper the authors investigate the prediction performance of DNN as a QSAR method and compare it with RF performance that is somehow considered as a gold standard in this field.
Motivation
At the first stage of drug discovery there are a huge number of candidate compounds that can be combined to produce a new drug. This process may involve a large number of compounds (>100 000) and a large number of descriptors (several thousands) that have different biological activity. Predicting all biological activities for all compounds need a lot number of experiments. The in silico discovery and using the optimization algorithms can substantially reduce the experiment work that need to be done. It was hypothesized that DNN models outperform RF models.
Methods
In order to compare the prediction performance of methods, DNN and RF fitted to 15 data sets from a pharmaceutical company, Merck. The smallest data set has 2092 molecules with 4596 unique AP, DP descriptors. Each molecule is represented by a list of features, i.e. “descriptors” in QSAR nomenclature. The descriptors are substructure descriptors (e.g., atom pairs (AP), MACCS keys, circular fingerprints, etc.) and donor-descriptors (DP). Both descriptors are of the following form:
atom type i − (distance in bonds) − atom type j
Where for AP, atom type includes the element, number of nonhydrogen neighbors, and number of pi electrons. For DP, atom type is one of seven (cation, anion, neutral donor, neutral acceptor, polar, hydrophobe, and other). A separate group of 15 different data sets as Additional Data Sets were used to validate the conclusions acquired from the Kaggle data sets. Each of these data sets was split into train and test set. The metric to evaluate prediction performance of methods is coefficient of determination ([math]\displaystyle{ R^2 }[/math]).
To run a RF, 100 trees were generated with m/3 descriptors used at each branch-point, where m was the number of unique descriptors in the training set. The tree nodes with 5 or fewer molecules were not split further. The trees parallelized to run one tree per processor on a cluster to run larger data sets in a reasonable time.
The DNNs with input descriptors X of a molecule and output of the form [math]\displaystyle{ O=f(\sum_{i=1}^{N} w_ix_i+b) }[/math] were fitted to data sets. Since many different parameters, such as number of layers, neurons, influence the performance of a deep neural net, Ma and his colleagues did a sensitivity analysis. They trained 71 DNNs with different parameters for each set of data. the parameters that they were considered were parameters related to:
-Data (descriptor transformation: no transformation, logarithmic transformation, or binary transformation.
-Network architecture: number of hidden layers, number of neurons in each hidden layer.
-Activation functions: sigmoid or rectified linear unit.
-The DNN training strategy: single training set or joint from multiple sets, percentage of neurons to drop-out in each layer.
-The mini-batched stochastic gradient descent procedure in the BP algorithm: the minibatch size, number of epochs
-Control the gradient descent optimization procedure: learning rate, momentum strength, and weight cost strength.
In addition to the effect of these parameters on the DNN, the authors were interested in evaluating consistency of results for a diverse set of QSAR tasks. Due to time-consuming process of evaluating the effect of the large number of adjustable parameters, a reasonable number of parameter settings were selected by adjusting the values of one or two parameters at a time, and then calculate the [math]\displaystyle{ R^2 }[/math] for DNNs trained with the selected parameter settings. These results allowed them to focus on a smaller number of parameters, and to finally generate a set of recommended values for all algorithmic parameters, which can lead to consistently good predictions.
Results
For the first object of this paper that was comparing the performance of DNNs to Rf, over over 50 DNNs were trained using different parameter settings. These parameter settings were arbitrarily selected, but they attempted to cover a sufficient range of values for each adjustable parameter. Figure 1 shows the difference in [math]\displaystyle{ R^2 }[/math] between DNNs and RF for each kaggle data set. Each column represents a QSAR data set, and each circle represents the improvement of a DNN over RF.
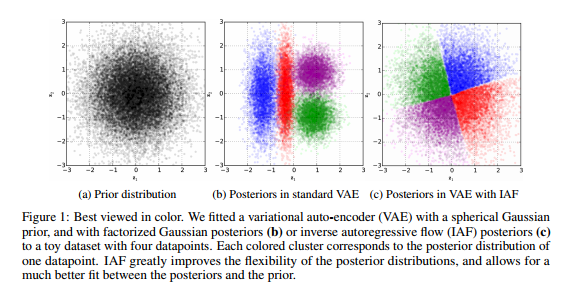
comparing the performance of different models shows that even when the worst DNN parameter setting was used for each QSAR task, the average R2 would be degraded only from 0.423 to 0.412, merely a 2.6% reduction. These results suggest that DNNs can generally outperform RF( table below).
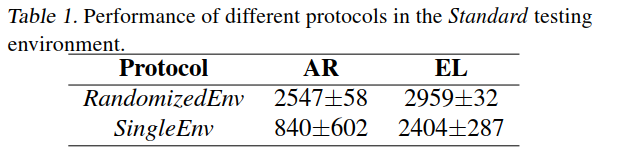
The difference in [math]\displaystyle{ R^2 }[/math] between DNN and RF by changing the the network architecture is shown in Figure 2. In order to limit the number of different parameter combinations they fixed the number of neurons in each hidden layer. Thirty two DNNs were trained for each data set by varying number of hidden layers and number of neurons in each layer while the other key adjustable parameters were kept unchanged. It is seen that when the number of hidden layers are two, having a small number of neurons in the layers degrade the predictive capability of DNNs. It can also be seen that, given any number of hidden layers, once the number of neurons per layer is sufficiently large, increasing the number of neurons further has only a marginal benefit. In Figure 2 we can see that the neural network with only one hidden layer and 12 neurons in each layer achieved the same average predictive capability as RF . This size of neural network is indeed comparable with that of the classical neural network used in QSAR.
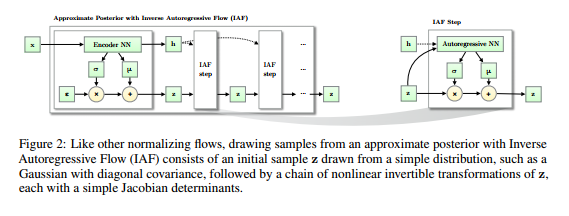
To decide which activation function, Sigmoid or ReLU, performs better, at least 15 pairs of DNNs were trained for each data set. Each pair of DNNs shared the same adjustable parameter settings, except that one DNN used ReLU as the activation function, while the other used Sigmoid function. The data sets where ReLU is significantly better than Sigmoid are colored in blue, and marked at the bottom with “+”s. The difference was tested by one-sample Wilcoxon test. In contrast, the data set where Sigmoid is significantly better than ReLU is colored in black, and marked at the bottom with “−”s( Figure 3). In 53.3% (8 out of 15) data sets, ReLU is statistically significantly better than Sigmoid. Overall ReLU improves the average [math]\displaystyle{ R^2 }[/math] over Sigmoid by 0.016.
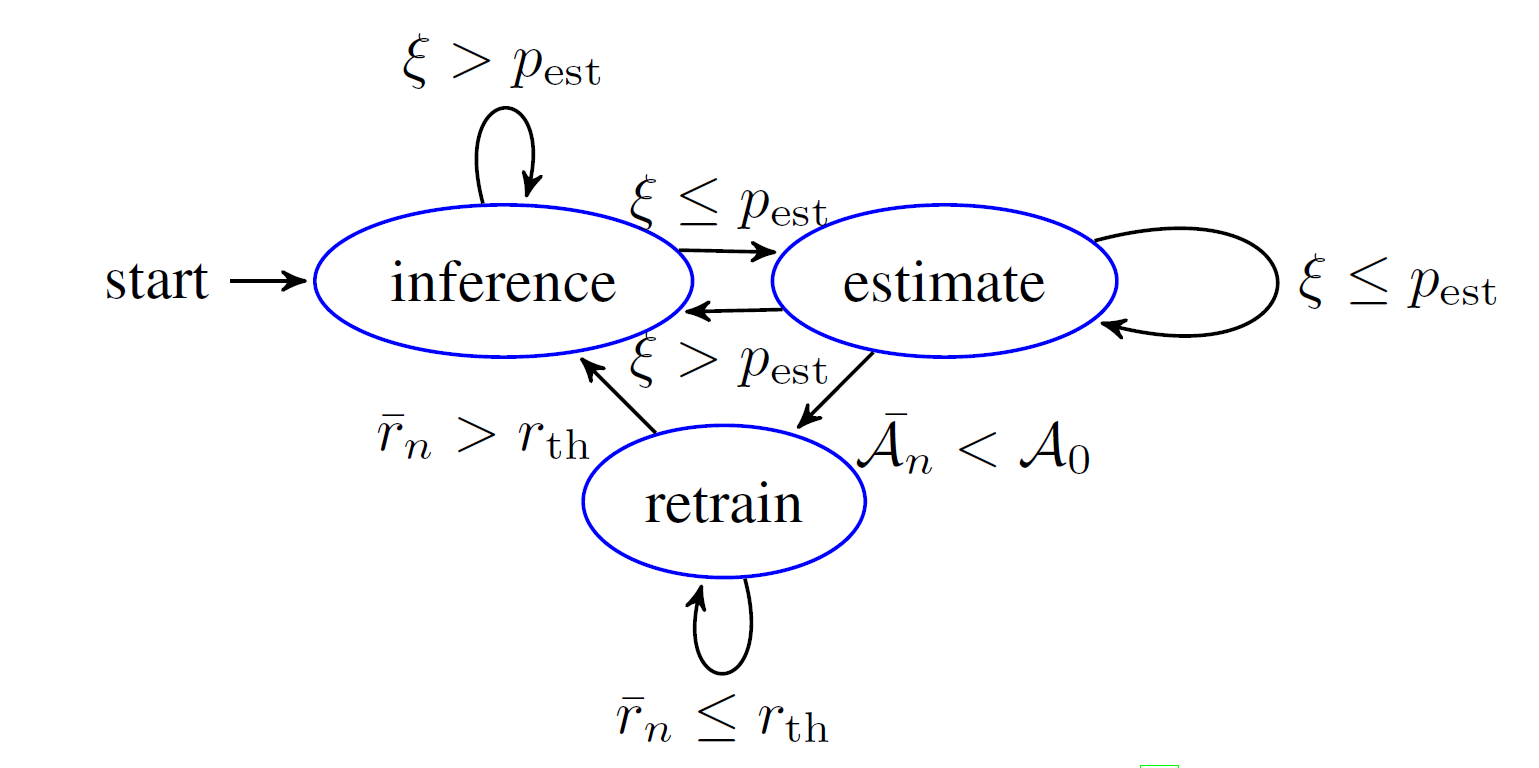
Figure 4 presents the difference between joint DNNs trained with multiple data sets and the individual DNNs trained with single data sets. Average over all data sets, there seems to joint DNN has a better performance rather single training. However, the size of the training sets plays a critical role on whether a joint DNN is beneficial. For the two very largest data sets (i.e., 3A4 and LOGD), the individual DNNs seem better, indicating that joint DNNs are more proper for not much large data sets.
The authors refine their selection of DNN adjustable parameters by studying the results of previous runs. They used the logarithmic transformation, two hidden layers, at least 250 hidden layers an activation function of ReLU. The results are shown in Figure 5. Comparison of these results with those in Figure 1 indicates that now there are 9 out of 15 data sets, whereDNNs outperforms RF even with the “worst” parameter setting, compared with 4 out of 15. The [math]\displaystyle{ R^2 }[/math] averaged over all DNNs and all 15 data sets is 0.051 higher than that of RF.
as a conclusion for the sensitivity analysis which had been done in this work, the authors gave a recommendation on the adjustable parameters of DNNs as below: -logarithmic transformation.
-four hidden layers, with number of neurons to be 4000, 2000, 1000, and 1000, respectively.
-The dropout rates of 0 in the input layer, 25% in the first 3 hidden layer, and 10% in the last hidden layer.
-The activation function of ReLU.
-No unsupervised pretraining. The network parameters should be initialized as random values.
-Large number of epochs.
-Learning rate of 0.05, momentum strength of 0.9, and weight cost strength of 0.0001.
To check the consistency of DNNs predictions as was one of concerns of authors, they compared the performance of RF with DNN on 15 additional QSAR data sets. Each additional data set was time-split into training and test sets in the same way as the Kaggle data sets. Individual DNNs were trained from the training set using the recommended parameters.[math]\displaystyle{ R^2 }[/math] of the DNN and RF were calculated from the test sets. Table below presents the results for the additional data sets. It is seen that the DNN with recommended parameters outperforms RF in 13 out of the 15 additional data sets. The mean [math]\displaystyle{ R^2 }[/math] of DNNs is 0.411, while that of RFs is 0.361, which is an improvement of 13.9%.
Both RF and DNN can be efficiently speeded up using high performance computing technologies, but in a different way due to the inherent difference in their algorithms.
Discussion
This paper demonstrate that DNN in most cases can be used as a practical QSAR method in place of RF which is now as a gold standard in the field of drug discovery. Although, the magnitude of the change in coefficient of determination relative to RF is small in some data sets, on average its better than RF. The paper recommends a set of values for all DNN algorithmic parameters, which are appropriate for large QSAR data sets in an industrial drug discovery environment. The authors gave some recommendation about how RF and DNN can be efficiently sped up using high performance computing technologies. They suggest that RF can be accelerated using coarse parallelization on a cluster by giving one tree per node. In contrast, DNN can efficiently make use of the parallel computation capability of a modern GPU.
Future Works
In opposite of our expectation that unsupervised pretraining plays a critical role in the success of DNNs, in this study it had an inverse effect on the performance of QSAR tasks which need to be worked. Although the paper had some recommendations about the adjustable parameters of DNNs, there is still need to develop an effective and efficient strategy for refining these parameters for each particular QSAR task. The result of current paper suggested that cross-validation failed to be effective for fine-tuning the algorithmic parameters. Therefore, instead of using automatic methods for tuning DNN parameters, new approaches that can better indicate a DNN’s predictive capability in a time-split test set are needed to be developed.
Bibliography
<references />