Summary for survey of neural networked-based cancer prediction models from microarray data
Presented by
Rao Fu, Siqi Li, Yuqin Fang, Zeping Zhou
Introduction
Microarray technology can help researchers quickly detect genetic information and is widely used to analyze genetic diseases. Researchers use this technology to compare normal and abnormal cancerous tissues to gain insights into cancer pathology.
However, due to the high dimensionality of the gene expressions, the model's accuracy and computation time might be affected. The following two approaches are adopted to cope with this problem: the feature selection method and the feature creating method. Like the well-known principal component analysis method, the former one aims to focus on key features and ignore minor noises. On the other hand, the latter one, similar to scale-invariant feature transformations, aims to combine existing features or map them to a new low-dimensional space.
Compared to others' neural network models, this paper is specifically designed for predicting cancer using gene expression data. Thus, we will review the latest neural network-based cancer prediction models by presenting the methodology of preprocessing, filtering, prediction, and clustering gene expressions.
Background
Neural Network
Neural networks are often used to solve non-linear complex problems. It is an operational model consisting of a large number of neurons connected to each other by different weights. In this network structure, each neuron is related to an activation function such as sigmoid or rectified linear activation functions. To train the network, the inputs are fed forward and the activation function value is calculated at every neuron. The difference between the output of the neural network and the actual value is what we call an error.
The backpropagation mechanism is one of the most commonly used algorithms in solving neural network problems. By using this algorithm, we optimize the objective function by propagating back the generated error through the network to adjust the weights.
In the next sections, we will use the above algorithm but with different network architectures and a different number of neurons to review the neural network-based cancer prediction models for learning the gene expression features.
Cancer prediction models
Cancer prediction models often contain more than 1 method to achieve high prediction accuracy with a more accurate prognosis and it also aims to reduce the cost of patients.
High dimensionality and spatial structure are the two main factors that can affect the accuracy of the cancer prediction models. They add irrelevant noisy features to our selected models. We have 3 ways to determine the accuracy of a model.
The first way is called the ROC curve. ROC curves, receiver operating characteristic curves, are graphs that show the true positive rate against the false-positive rate [4]. It reflects the sensitivity of the response to the same signal stimulus under different criteria. To test its validity, we need to consider it with the confidence interval. Usually, a model is considered acceptable when its ROC is greater than 0.7.
A different machine learning problem is predicting the survival time. The performance of a model that predicts survival time can be measured using two metrics. The problem of predicting survival time can be seen as a ranking problem, where survival times of different subjects are ranked against each other. CI (Concordance Index) is a measure of how good a model ranks survival times and explains the concordance probability of the predicted and observed survival. The closer its value to 0.7, the better the model is. We can express the ordering of survival times in an order graph [math]\displaystyle{ G = (V, E) }[/math] where the vertices [math]\displaystyle{ V }[/math] are the individual survival times, and the edges [math]\displaystyle{ E_{ij} }[/math] from individual [math]\displaystyle{ i }[/math] to [math]\displaystyle{ j }[/math] indicate that [math]\displaystyle{ T_i \lt T_j }[/math], where [math]\displaystyle{ T_i, T_j }[/math] are the survival times for individuals [math]\displaystyle{ i,j }[/math] respectively. Then we can write [math]\displaystyle{ CI = \frac{1}{|E|}\sum_{i,j}I(f(i) \lt f(j)) }[/math] where [math]\displaystyle{ |E| }[/math] is the number of edges in the order graph (ie. the total number of comparable pairs), and [math]\displaystyle{ I(f(i) \lt f(j)) = 1 }[/math] if the predictor [math]\displaystyle{ f }[/math] correctly ranks [math]\displaystyle{ T_i \lt T_j }[/math] [3].
Another metric is the Brier score, which measures the average difference between the observed and the estimated survival rate in a given period of time. It ranges from 0 to 1, and a lower score indicates higher accuracy. It is defined as [math]\displaystyle{ \frac{1}{n}\sum_{i=1}^n(f_i - o_i)^2 }[/math] where [math]\displaystyle{ f_i }[/math] is the predicted survival rate, and [math]\displaystyle{ o_i }[/math] is the observed survival rate [2].
Neural network-based cancer prediction models
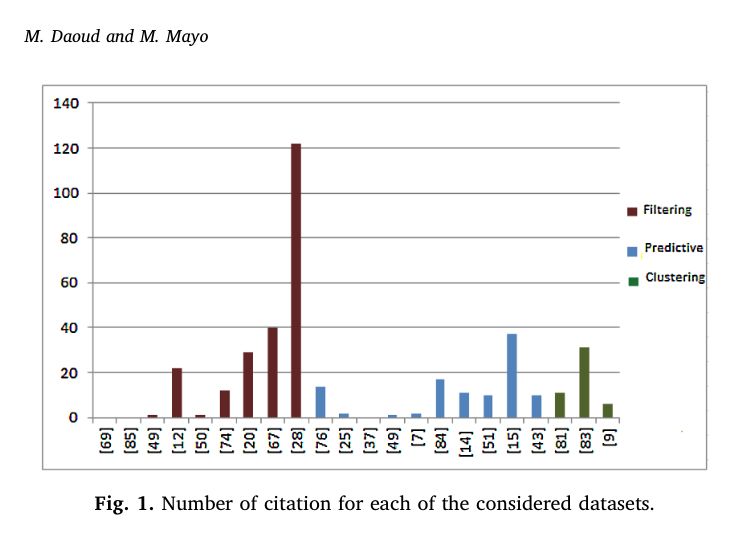
An extensive search relevant to neural network-based cancer prediction was performed using Google scholar and other electronic databases namely PubMed and Scopus with keywords such as “Neural Networks AND Cancer Prediction” and “gene expression clustering”, and only articles between 2013 and 2018 with available accessibility were considered. The chosen papers covered cancer classification, discovery, survivability prediction, and statistical analysis models. The following figure 1 shows a graph representing the number of citations including filtering, predictive, and clustering for chosen papers.
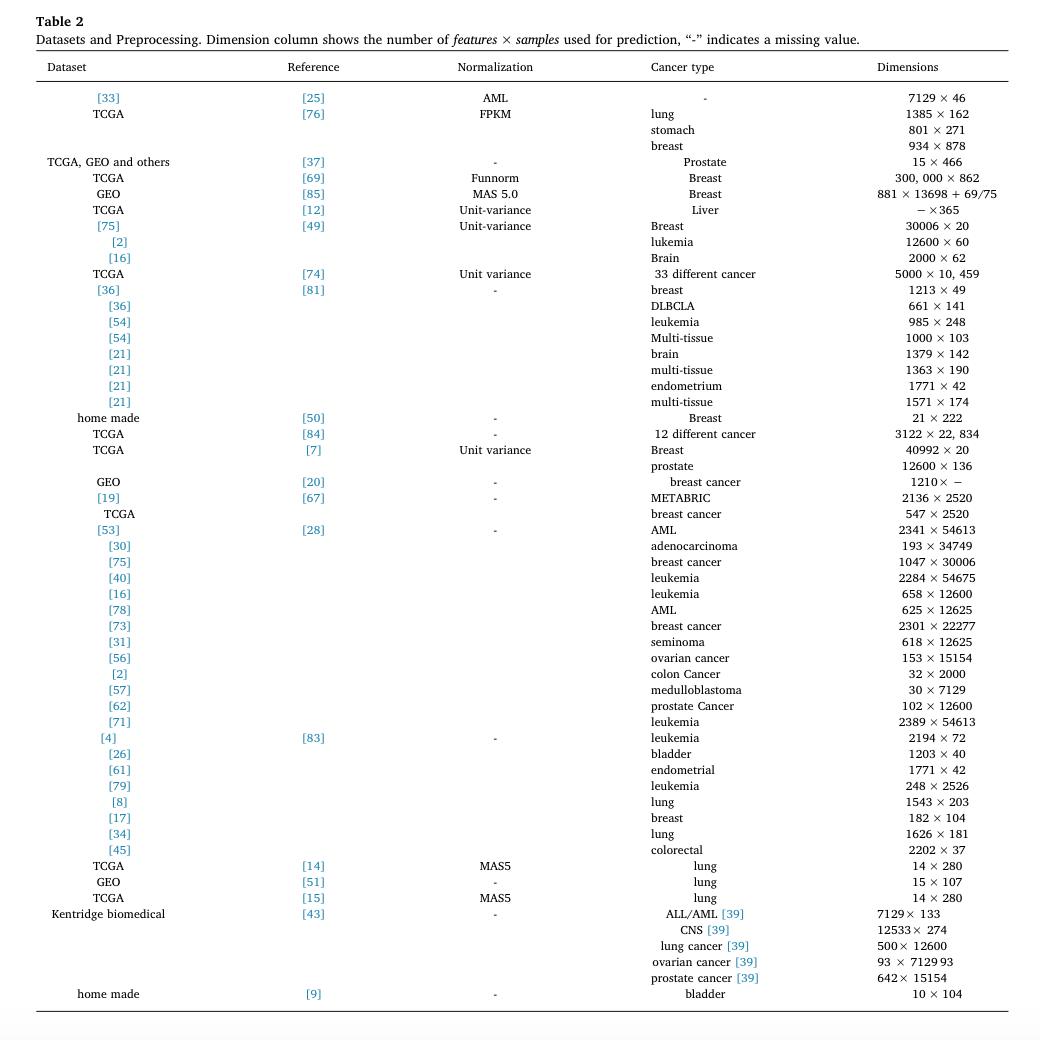
Datasets and preprocessing
Most studies investigating automatic cancer prediction and clustering used datasets such as the TCGA, UCI, NCBI Gene Expression Omnibus and Kentridge biomedical databases. There are a few techniques used in processing dataset including removing the genes that have zero expression across all samples, Normalization, filtering with p-value > [math]\displaystyle{ 10^{-5} }[/math] to remove some unwanted technical variation and [math]\displaystyle{ \log_2 }[/math] transformations. Statistical methods, neural networks, were applied to reduce the dimensionality of the gene expressions by selecting a subset of genes. Principle Component Analysis (PCA) can also be used as an initial preprocessing step to extract the dataset's features. The PCA method linearly transforms the dataset features into lower dimensional space without capturing the complex relationships between the features. However, simply removing the genes that were not measured by the other datasets could not overcome the class imbalance problem. In that case, one research used Synthetic Minority Class Over Sampling method to generate synthetic minority class samples, which may lead to a sparse matrix problem. This is because variability needed to be included in the generated samples. Consequently, randomly chosen features would be set to zero, which may cause the sparse matrix problem (Daoud & Mayo, 2019). Clustering was also applied in some studies for labeling data by grouping the samples into high-risk, low-risk groups, and so on.
The following table presents the dataset used by considered reference, the applied normalization technique, the cancer type and the dimensionality of the datasets.
Neural network architecture
Most recent studies reveal that neural network methods are used for filtering, predicting, and clustering in cancer prediction.
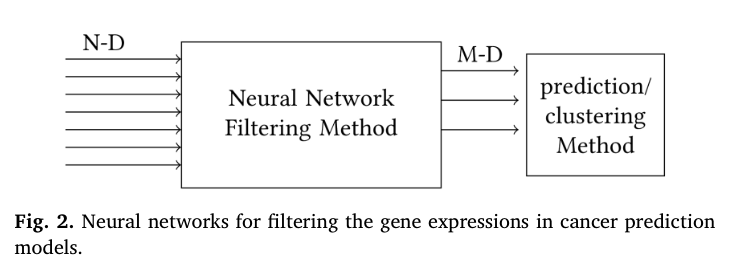
- filtering: Filter the gene expressions to eliminate noise or reduce dimensionality. Then use the resulted features with statistical methods or machine learning classification and clustering tools as figure 2 indicates.
- predicting: Extract features and improve the accuracy of prediction (classification).
- clustering: Divide the gene expressions or samples based on similarity.
All the neurons in the neural network work together as feature detectors to learn the features from the input. In order to categorize a neural network as filtering, predicting, or clustering method, we looked at the overall role that network provided within the framework of cancer prediction. Filtering methods are trained to remove the input’s noise and to extract the most representative features that best describe the unlabeled gene expressions. Predicting methods are trained to extract the features that are significant to prediction, therefore its objective functions measure how accurately the network is able to predict the class of input. Clustering methods are trained to divide unlabeled samples into groups based on their similarities.
Building neural networks-based approaches for gene expression prediction
According to our survey, the representative codes are generated by filtering methods with dimensionality M that is smaller or equal to N, where N is the dimensionality of the input. Some other machine learning algorithms such as naïve Bayes or k-means can be used together with the filtering.
Predictive neural networks are supervised, which find the best classification accuracy; meanwhile, clustering methods are unsupervised, which group similar samples or genes together.
The goal of training prediction is to enhance the classification capability, and the goal of training classification is to find the optimal group for a new test set with unknown labels.
Neural network filters for cancer prediction
In the preprocessing step to classification, clustering, and statistical analysis, the autoencoders are more and more commonly-used, to extract generic genomic features. An autoencoder is composed of the encoder part and the decoder part. The encoder part is to learn the mapping between high-dimensional unlabeled input I(x) and the low-dimensional representations in the middle layer(s), and the decoder part is to learn the mapping from the middle layer’s representation to the high-dimensional output O(x). The reconstruction of the input can take the Root Mean Squared Error (RMSE) or the Logloss function as the objective function.
$$ RMSE = \sqrt{ \frac{\sum{(I(x)-O(x))^2}}{n} } $$
$$ Logloss = \sum{(I(x)\log(O(x)) + (1 - I(x))\log(1 - O(x)))} $$
There are several types of autoencoders, such as stacked denoising autoencoders, contractive autoencoders, sparse autoencoders, regularized autoencoders, and variational autoencoders. The architecture of the networks varies in many parameters, such as depth and loss function. Each example of an autoencoder mentioned above has a different number of hidden layers, different activation functions (e.g. sigmoid function, exponential linear unit function), and different optimization algorithms (e.g. stochastic gradient descent optimization, Adam optimizer).
Overfitting is a major problem that most autoencoders need to deal with to achieve high efficiency of the extracted features. Regularization, dropout, and sparsity are common solutions.
The neural network filtering methods were used by different statistical methods and classifiers. The conventional methods include Cox regression model analysis, Support Vector Machine (SVM), K-means clustering, t-SNE and so on. The classifiers could be SVM or AdaBoost or others.
By using neural network filtering methods, the model can be trained to learn low-dimensional representations, remove noises from the input, and gain better generalization performance by re-training the classifier with the newest output layer.
Neural network prediction methods for cancer
The prediction based on neural networks can build a network that maps the input features to an output with a number of neurons, which could be one or two for binary classification or more for multi-class classification. It can also build several independent binary neural networks for the multi-class classification, where the technique called “one-hot encoding” is applied.
The codeword is a binary string [math]\displaystyle{ C'k }[/math] of length k whose j’th position is set to 1 for the j’th class, while other positions remain 0. The process of the neural networks is to map the input to the codeword iteratively, whose objective function is minimized in each iteration.
Such cancer classifiers were applied to identify cancerous/non-cancerous samples, a specific cancer type, or the survivability risk. MLP models were used to predict the survival risk of lung cancer patients with several gene expressions as input. The deep generative model DeepCancer, the RBM-SVM and RBM-logistic regression models, the convolutional feedforward model DeepGene, Extreme Learning Machines (ELM), the one-dimensional convolutional framework model SE1DCNN, and GA-ANN model are all used for solving cancer issues mentioned above. This paper indicates that the performance of neural networks with MLP architecture as a classifier is better than those of SVM, logistic regression, naïve Bayes, classification trees, and KNN.
Neural network clustering methods in cancer prediction
Neural network clustering belongs to unsupervised learning. The input data are divided into different groups according to their feature similarity.
The single-layered neural network SOM, which is unsupervised and without a backpropagation mechanism, is one of the traditional model-based techniques to be applied to gene expression data. The measurement of its accuracy could be Rand Index (RI), which can be improved to Adjusted Random Index (ARI) and Normalized Mutation Information (NMI).
$$ RI=\frac{TP+TN}{TP+TN+FP+FN}$$
In general, gene expression clustering considers either the relevance of samples-to-cluster assignment or that of gene-to-cluster assignment or both. The high dimensionality of gene expression samples poses a problem for traditional clustering algorithms such as k-means clustering, which uses a distance function to separate samples. Such an approach is not viable for high dimensional datasets. To solve the high dimensionality problem, there are two methods: clustering ensembles by running a single clustering algorithm several times, each of which has different initialization or number of parameters; and projective clustering by only considering a subset of the original features.
SOM was applied on discriminating future tumor behavior using molecular alterations, whose results were not easy to be obtained by classic statistical models. Then this paper introduces two ensemble clustering frameworks: Random Double Clustering-based Cluster Ensembles (RDCCE) and Random Double Clustering-based Fuzzy Cluster Ensembles (RDCFCE). Their accuracies are high, but they have not taken the gene-to-cluster assignment into consideration.
Also, the paper provides a double SOM based Clustering Ensemble Approach (SOM2CE) and double NG-based Clustering Ensemble Approach (NG2CE), which are robust to noisy genes. Moreover, Projective Clustering Ensemble (PCE) combines the advantages of both projective clustering and ensemble clustering, which is better than SOM and RDCFCE when there are irrelevant genes.
Summary
Cancer is a disease with a high mortality rate that kills millions of people every year, and it’s essential to analyze gene expression for discovering gene abnormalities and increasing survivability as a consequence. The previous analysis in the paper reveals that neural networks are essentially used for filtering the gene expressions, predicting their class, or clustering them.
Neural network filtering methods are used to reduce the dimensionality of the gene expressions and remove their noise. In the article, the authors recommended deep architectures in comparison to shallow architectures for best practice, as they combine many nonlinearities.
Neural network prediction methods can be used for both binary and multi-class problems. In the binary case, the network architecture has only one or two output neurons that diagnose a given sample as cancerous or non-cancerous. In comparison, the number of the output neurons in multi-class problems is equal to the number of classes. The authors suggested that the deep architecture with convolution layers which was the most recently used model proved efficient capability in predicting cancer subtypes, as it captures the spatial correlations between gene expressions. Clustering is another analysis tool that is used to divide the gene expressions into groups. The authors indicated that a hybrid approach combining both the assembling, clustering and projective clustering is more accurate than using single-point clustering algorithms, such as SOM, since those methods do not have the capability to distinguish the noisy genes.
Discussion
There are some technical problems that can be considered and improved for building new models.
1. Overfitting: Since gene expression datasets are high dimensional and have a relatively small number of samples, it would be likely to properly fits the training data but not accurate for test samples due to the lack of generalization capability. The ways to avoid overfitting can be: (1). adding weight penalties using regularization; (2). using the average predictions from many models trained on different datasets; (3). dropout. (4) Augmentation of the dataset to produce more "observations".
2. Model configuration and training: In order to reduce both the computational and memory expenses but also with high prediction accuracy, it’s crucial to properly set the network parameters. The possible ways can be: (1). proper initialization; (2). pruning the unimportant connections by removing the zero-valued neurons; (3). using ensemble learning framework by training different models using different parameter settings or using different parts of the dataset for each base model; (4). Using SMOTE for dealing with class imbalance on the high dimensional level.
3. Model evaluation: In Braga-Neto and Dougherty's research, they have investigated several model evaluation methods: cross-validation, substitution and bootstrap methods. The cross-validation was found to be unreliable for small size data since it displayed excessive variance. The bootstrap method proved more accurate predictability.
4. Study producibility: A study needs to be reproducible to enhance research reliability so that others can replicate the results using the same algorithms, data and methodology. Hence, the query used for getting the dataset should be stated.
Conclusion
This paper reviewed the most recent neural network-based cancer prediction models and gene expression analysis tools. The analysis indicates that the neural network methods are able to serve as filters, predictors, and clustering methods, and also showed that the role of the neural network determines its general architecture. The authors showed that Neural Network filtering methods are a way of reducing the dimensionality of the gene expressions, as well as removing their noise for better model fitting. To give suggestions for future neural network-based approaches, the authors highlighted some critical points that have to be considered such as overfitting and class imbalance, and suggest choosing different network parameters or combining two or more of the presented approaches. One of the biggest challenges for cancer prediction modelers is deciding on the network architecture (i.e. the number of hidden layers and neurons), as there are currently no guidelines to follow to obtain high prediction accuracy. The authors discovered that there is no algorithm available to concretely determine an optimal number of hidden layers or nodes and found that many papers simply implemented a trial and error method to reduce loss in the model.
Critiques
While results indicate that the functionality of the neural network determines its general architecture, the decision on the number of hidden layers, neurons, hypermeters, and learning algorithms is made using trial-and-error techniques. Therefore improvements in this area of the model might need to be explored in order to obtain better results and in order to make more convincing statements.
An issue that one must be mindful of is the underlying distribution of data. Cancer is an extremely complex genetic disease and the predictions would depend on so many variables, a number of which will not even be present in the dataset as they might have been collected. So there is a need for extensive validation when it comes to applying deep learning methods to cancer-related data.
In the field of medical sciences and molecular biology, interpretability of results is imperative as often experts seek not just to solve the issue at hand but to understand the causal relationships. Having a high ROC value may not necessarily convince other experts on the validity of the finding because the underlying details of cancer symptoms have been abstracted in a complex neural network as a black box. However, the neural network clustering method suggested in this paper does offer a good compromise because it enables humans to visual low-level features but still gives experts the control on making various predictions using well-studied traditional techniques.
With high dimensionality features, kernel SVM is another option for cancer prediction. Jiang et. al. developed a Hadamard Kernel for predicting breast cancer using gene expression data, and it utilizes the Kernel trick to avoid high computational efforts (link: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5763304/). Compared against linear, quadratic, RBF and correlation kernels, Hadamard Kernel performs best with the highest averaged area under the ROC curve (AUC) value. It may be interesting to compare the performance and accuracy between the Hadamard Kernel and cancer prediction models with various number of hidden layers and neurons.
Although the authors presented technical details about data processing, training approaches evaluation metric, and addressed many practical issues that can be considered for cancer prediction, no novel methods or models are proposed. It's more like a proof-of-concept about the feasibility of different models on cancer prediction.
It would be interesting for the researchers to compare the performance between causal inference and neural network models on this data.
As the authors indicate neural networks would be a useful tool for cancer prediction models, the article is lacking an example for implementing neural networks to provide persuasive support for their arguments.
The inheritance of cancer is complex and changeable. The predicted variables are therefore very complicated, so for the model of the learning data set, a more adequate training set is needed to learn. And multi-party verification of the learned model.
The authors mentioned many different neural network models and compared them. It would be better if more details of a commonly applied model with relatively high accuracy could be given such as how the model is built. An article named Convolutional neural network models for cancer type prediction based on gene expression gives explanations of CNN in detail.
The authors briefly discussed methods and algorithms being used in the presented paper in their summary. However, a very little amount of technical details were provided to the readers. The summary itself is lacking specific examples for the aforementioned algorithms, and datasets which were used in the original analysis were only introduced in one or two sentences. As a result, the summary and conclusion appear to be unconvincing to the readers.
PCA can still be used as an initial preprocessing step even if it is used in a neural network whose data dimension is reduced. By merging the PCA component with a random number of original functions, some good techniques can be adopted to enable the network to capture more useful relationships. Correlation based feature selection (CFS) might be also applied in the data pre-processing step to reduce the dimensionality.
The key part of this model is to extract the features from the model. However, cancer may depends on many explanatory variables. Thus how do we know which feature should we extract in the data preprocessing. Since there are correlations between each variables. Authors did not specify this situation.
From the biology side of view, gene expression is really complicated. Thus reducing dimension may or may not be the best way of predicting cancer, and this should be a controversial topic.
The author should explore more on the reason predicting certain cancers are more accurate than other cancers. Also the medical conditions are different among all individuals, there are more features that need to be considered when doing this model.
Reference
[1] Daoud, M., & Mayo, M. (2019). A survey of neural network-based cancer prediction models from microarray data. Artificial Intelligence in Medicine, 97, 204–214.
[2] Brier GW. 1950. Verification of forecasts expressed in terms of probabilities. Monthly Weather Review 78: 1–3
[3] Harald Steck, Balaji Krishnapuram, Cary Dehing-oberije, Philippe Lambin, Vikas C. Raykar. On ranking in survival analysis: Bounds on the concordance index. In Advances in Neural Information Processing Systems (2008), pp. 1209-1216
[4] Google Developers. (2020 February 10). Classification: ROC Curve and AUC. Machine Learning Crash Course. https://developers.google.com/machine-learning/crash-course/classification/roc-and-auc
[5] Mostavi, M., Chiu, YC., Huang, Y. et al. Convolutional neural network models for cancer type prediction based on gene expression. BMC Med Genomics 13, 44 (2020). https://doi.org/10.1186/s12920-020-0677-2