relevant Component Analysis
First paper: Shental et al., 2002 <ref>N. Shental, T. Hertz, D. Weinshall, and M. Pavel, "Adjustment Learning and Relevant Component Analysis," Proc. European Conference on Computer Vision (ECCV), 2002, pp. 776-790.</ref>
Irrelevant data variability often causes difficulties in classification and clustering tasks. For example, when data variability is dominated by environment conditions, such as global illumination, nearest-neighbour classification in the original feature space may be very unreliable. The goal of Relevant Component Analysis (RCA) is to find a transformation that amplifies relevant variability and suppresses irrelevant variability.
- Definition of irrelevant variability: We say that data variability is correlated with a specific task "if the removal of this variability from the data deteriorates (on average) the results of clustering or retrieval" [1]. Variability is irrelevant if it is "maintained in the data" but "not correlated with the specific task" [1].
To achieve this goal, Shental et al. introduced the idea of chunklets – "small sets of data points, in which the class label is constant, but unknown" [1]. As we will see, chunklets allow irrelevant variability to be suppressed without needing fully labelled training data. Since the data come unlabelled, the chunklets "must be defined naturally by the data": for example, in speaker identification, "short utterances of speech are likely to come from a single speaker" [1]. The authors coin the term adjustment learning to describe learning using chunklets; adjustment learning can be viewed as falling somewhere between unsupervised learning and supervised learning.
Relevant Component Analysis tries to find a linear transformation W of the feature space such that the effect of irrelevant variability is reduced in the transformed space. That is, we wish to rescale the feature space and reduce the weights of irrelevant directions. The main premise of RCA is that we can reduce irrelevant variability by reducing the within-class variability. Intuitively, a direction which exhibits high variability among samples of the same class is unlikely to be useful for classification or clustering.
RCA assumes that the class covariances are all equal. If we allow this assumption, it makes sense to rescale the feature space using a whitening transformation based on the common class covariance Σ. This gives the familiar transformation W = VΛ-1/2, where V and Λ can be found by the singular value decomposition of Σ.
With labelled data estimating Σ is straightforward, but in RCA labelled data is not available and an approximation is calculated using chunklets. The chunklet scatter matrix is calculated by
- [math]\displaystyle{ S_{ch} = \frac{1}{|\Omega|}\sum_{n=1}^N|H_n|Cov(H_n) }[/math]
where |Ω| is the size of the data set, Hn is the nth chunklet, |Hn| is the size of the nth chunklet, and N is the number of chunklets.
Intuitively, this is a weighted average of the chunklet covariances, with weight proportional to the size of the chunklet. Here, we are seeking a chunklet which makes a good mean value approximation of a class, regardless of the chunklet’s size. However the size matters as any increasing in the size would increase the likelihood of well-done approximation of the class mean.
The steps of the RCA algorithm are as follows:
- "1. Calculate Sch... Let r denote its effective rank (the number of singular values of Sch which are significantly larger than 0).
- 2. Compute the total covariance (scatter) matrix of the original data ST, and project the data using PCA to its r largest dimensions.
- 3. Project Sch onto the reduced dimensional space, and compute the corresponding whitening transformation W.
- 4. Apply W to the original data (in the reduced space)." [1]
Those directions in which the data variability is due to class variability are irrelevant for classification and the computed W assigns lower weight to these directions.
Experimental Results: Face Recognition
The authors demonstrated the performance of RCA for the task of face recognition using the yaleA database. The database contains 155 face images of 15 people; lighting conditions and facial expression are varied across images. RCA is compared with the Eigenface method (based on PCA) and the Fisherface method (based on Fisher’s Linear Discriminant) for both nearest neighbour classification and clustering-based classification. In this dataset, the data is not naturally divided into chunklets, so the authors randomly sample chunklets given the ground-truth class (for example, if an individual is represented in 10 images, two chunklets may be formed by randomly partitioning the images into two groups of 5 images.)
For nearest neighbour classification, RCA outperforms Eigenface but does slightly worse than Fisherface. For clustering, RCA performs better than Eigenface and comparably to Fisherface. The authors pointed out that these experimental results are encouraging as Fisherface is a supervised method.
In <ref> M. Sorci,G. Antonini, and Jean-Philippe Thiran, "Fisher's discriminant and relevant component analysis for static facial expression classification."</ref>, it's shown that RCA in combination with FLD results in better classifier in the context of facial expression recognition framework as compared to RCA alone. This combination has results comparable to SVM.
Experimental Results: Surveillance
In a second experiment, the authors used surveillance video footage divided into discrete clips in which a single person is featured. The same person can appear in multiple clips, and the task was to retrieve all clips in which a query person appears. A colour histogram is used to represent a person. Sources of irrelevant variation include reflections, occlusions, and illumination. In this experiment, the data does come naturally in chunklets: each clip features a single person, so frames in the same clip from a chunklet. Figure 7 in the paper shows the results of k-nearest neighbour classification (not reproduced here for copyright reasons).
Adjustment Learning
Adjusting learning is a learning paradigm the authors coined in their paper. As will be explained below, adjustment learning lies between supervised and unsupervised learning.
In the supervised learning paradigm, each data point is given with an associated label; the system learns the mapping from the set of data to the set of labels.
In the unsupervised learning paradigm, each data point is given without an associated label; the system aims to summarize how the data is organized and explain features of the data.
In the adjustment learning paradigm, each data point is associated with a label but the label is unknown. Moreover, the adjustment learning paradigm assumes that input data is naturally partitioned into small subsets, or chunklets, which are in turn subsets of equivalence classes that partition the input data. In other words, all data point in the same chunklet have the same (unknown) label; and different chunklets can be associated to the same label. The intuition behind adjust learning is that, by exploiting within-chunklet similarities, learning performance can be improved compared to unsupervised learning even when the labels are not given.
Adjustment learning aims to accomplish classification by exemplars, instead of by labels; meaning that an adjustment learning algorithm (e.g. RCA) has the capability to return data examples that "fall in the same class" with a given query datum. For example, the algorithm can return a set of Albert Einstein facial images when given an Albert Einstein facial image as the query datum. It is important to note that the above classification scheme does not involve explicitly labeling the data points (i.e. the algorithm does not (need to) label the images with "Albert Einstein").
Second Paper: Bar-Hillel et al., 2003 <ref> A. Bar-Hillel, T. Hertz, N. Shental, and D. Weinshall, "Learning Distance Functions using Equivalence Relations," Proc. International Conference on Machine Learning (ICML), 2003, pp. 11-18. </ref>
In a subsequent work [2], Bar-Hillel et al. described how RCA can be shown to optimize an information theoretic criterion, and compared the performance of RCA with the approach proposed by Xing et al. [3].
Information Maximization
According to information theory, "when an input X is transformed into a new representation Y, we should seek to maximize the mutual information I(X, Y) between X and Y under suitable constraints" [2]. In adjustment learning, we can think of the objective to be to keep chunklet points close to each other in the transformed space. More formally:
- [math]\displaystyle{ \max_{f \in F}I(X,Y) \quad s.t. \quad \frac{1}{p}\sum_{j=1}^k\sum_{i=1}^{n_j}||y_{ji} - m_j^y||^2 \le K }[/math]
where f is a transformation function, mjy is the mean of chunklet j in the transformed space, p is the total number of chunklet points, and K is a constant.
To maximize I(X,Y), we can simply maximize the entropy of Y, H(Y). This is because I(X,Y) = H(Y) – H(Y|X), and H(Y|X) is constant since the transformation is deterministic. Intuitively, since the transformation is deterministic there is no uncertainty in Y if X is known.
Now we would like to express H(Y) in terms of H(X). If the transformation is invertible, we have py(y) = px(x) / |J(x)|, where J(x) is the Jacobian of the transformation. Therefore,
- [math]\displaystyle{ \begin{align} H(Y) & = -\int_y p(y)\log p(y)\, dy \\ & = -\int_x p(x) \log \frac{p(x)}{|J(x)|} \, dx \\ & = H(X) + \langle \log |J(x)| \rangle_x \end{align} }[/math]
Assuming a linear transformation Y = AX, the Jacobian is simply equal to the constant |A|. So to maximize I(X,Y), we can maximize H(Y), and maximizing H(Y) amounts to maximizing |A|. Hence, the optimization objective can be updated as
- [math]\displaystyle{ \max_A |A| \quad s.t. \quad \frac{1}{p}\sum_{j=1}^k\sum_{i=1}^{n_j}||x_{ji} - m_j||^2_{A^tA} \le K }[/math]
This can also be expressed in terms of the Mahalanobis distance matrix B = AtA as follows, noting that log |A| = (1/2) log |B|.
- [math]\displaystyle{ \max_B |B| \quad s.t. \quad \frac{1}{p}\sum_{j=1}^k\sum_{i=1}^{n_j}||x_{ji} - m_j||^2_B \le K , \quad B \gt 0 }[/math]
The solution to this problem is [math]\displaystyle{ B = \tfrac{K}{N} \hat{C}^{-1} }[/math], where [math]\displaystyle{ \hat{C} }[/math] is the chunklet scatter matrix calculated in Step 1 of RCA. Thus, RCA gives the optimal Mahalanobis distance matrix up to a scale factor.
Within-Chunklet Distance Minimization
In addition, RCA minimizes the sum of within-chunklet squared distances. If we consider the optimization problem
- [math]\displaystyle{ \min_B \frac{1}{p}\sum_{j=1}^k\sum_{i=1}^{n_j}||x_{ji} - m_j||^2_B \quad s.t. \quad |B| \ge 1 }[/math]
then it can be shown that RCA once again gives the optimal Mahalanobis distance matrix up to a scale factor. This property suggests a natural comparison with Xing et al.’s method, which similarly learns a distance metric based on similarity side information. Xing et al.’s method assumes side information in the form of pairwise similarities and dissimilarities, and seeks to optimize
- [math]\displaystyle{ \min_B \sum_{(x_1,x_2) \in S} ||x_1 - x_2||^2_B \quad s.t. \sum_{(x_1,x_2) \in D} ||x_1 - x_2||_B \ge 1 , \quad B \ge 0 }[/math]
where S contains similar pairs and D contains dissimilar pairs. Comparing to the preceding optimization problem, if all chunklets have size 2 (i.e. the chunklets are just pairwise similarities), the objective function is the same up to a scale factor.
The authors compared the clustering performance of RCA with Xing et al.’s method <ref> E. Xing, A. Ng, M. Jordan, and S. Russell, "Distance metric learning with application to clustering with side-information", Advances in Neural Information Processing Systems, 2002. </ref> using six of the UC Irvine datasets. Clustering performance was measured using a normalized accuracy score defined as
- [math]\displaystyle{ \sum_{i \gt j}\frac{1 \lbrace 1 \lbrace c_i = c_j \rbrace = 1 \lbrace \hat{c}_i = \hat{c}_j \rbrace \rbrace}{0.5m(m-1)} }[/math]
where 1{ } is the indicator function, [math]\displaystyle{ \hat{c} }[/math] is the assigned cluster, and c is the true cluster. The score may be interpreted as the probability of correctly assigning two randomly drawn points.
Overall, RCA yielded an improvement over regular K-means and showed comparable performance to Xing et al.’s method, however RCA is more computationally efficient as it works with closed-form expressions while Xing et al.’s method requires iterative gradient descent.
Suggestions/Critique
- RCA makes effective use of limited side information in the form of chunklets, however in most applications the data does not naturally come in chunklets. Indeed, in the face recognition experiments, the authors had to make use of prior information to artificially create chunklets. It may be useful if the authors provided additional examples of applications where data is naturally partitioned into chunklets, to further motivate the applicability of RCA.
- RCA also assumes equal class covariances, which might limit its performance on many real-world datasets.
- In the UC Irvine experiments, RCA shows similar performance to Xing et al.’s method, but the authors noted that RCA is more computationally efficient. While they make a sensible logical argument (iterative gradient descent tends to be computationally expensive), providing experimental running times may help support and quantify this claim.
Why Equal Variances for Chanklets
In [2] authors suppose that [math]\displaystyle{ C_{m} }[/math] is the random variable which shows distribution of data in class [math]\displaystyle{ m }[/math] and then, assuming equality for class variances they calculate [math]\displaystyle{ S_{ch} }[/math] as it was mentioned above.
Further, suppose that data in class [math]\displaystyle{ m }[/math] are dependent on another source of variation [math]\displaystyle{ G }[/math] besides the class characteristics ([math]\displaystyle{ G }[/math] can be global variation or sensor characteristics). Now the random variable for [math]\displaystyle{ m }[/math]th class is [math]\displaystyle{ X=C_{m}+G }[/math], where global impact ([math]\displaystyle{ G }[/math]) is the same for all classes, [math]\displaystyle{ G }[/math] is independent of [math]\displaystyle{ C_{m} }[/math] and global variation is larger than class variation ([math]\displaystyle{ \Sigma_{m}\lt \Sigma_{G} }[/math]).
In this situation variance for class [math]\displaystyle{ m }[/math] will be [math]\displaystyle{ \Sigma_{m}+\Sigma_{G} }[/math], but by assumption it will be dominated by [math]\displaystyle{ \Sigma_{G} }[/math]. This result brings us back to the case [math]\displaystyle{ \Sigma_{m}=\Sigma_{G} }[/math] for all classes again.
Kernel RCA
Although RCA, computationally and technically, has significant advantages, there are some kind of situations for real problems that RCA fails to deal with them, i.e there are some restrictions along with RCA.
(i)- RCA only considers linear transformations and fails for nonlinear transformations (even for simple ones)
(ii)- since RCA acts in the input space, its number of parameters depends on the dimensionality of the feature vectors
(iii)- RCA requires the vectorial representation of data, which may not be possible for some kind of data to be naturally in this form; like protein sequences.
To overcome this restrictions Tesang and colleagues (2005)<ref> Tsang, I. W. and Colleagues; Kernel Relevant Component Analysis For Distance Metric Learning. International Joint Conference on Neural Networks, Montreal, Canada, July 31 - August 4, 2005 </ref> suggested to use kernel in RCA and showed how one can kernelize RCA.
Kernelizing RCA
For [math]\displaystyle{ k }[/math] given chunklets, each containing [math]\displaystyle{ n_{i} }[/math] patterns [math]\displaystyle{ \left\{x_{i,1},...,x_{i,n_{i}} \right\} }[/math] the covariance matrix of centered patterns is as follow:
[math]\displaystyle{ C=\frac{1}{n}\sum_{i=1}^{k}\sum_{j=1}^{n_{i}} \left(x_{i,j}-\bar{x}_{i}\right)\left(x_{i,j}-\bar{x}_{i}\right)^{'} }[/math]
and the associated whitening transform is as
[math]\displaystyle{ x\stackrel{}{\rightarrow}C^{-\frac{1}{2}}x }[/math]
Now let [math]\displaystyle{ \left\{x_{1,1},x_{1,2},...,x_{1,n_{1}},...,x_{k,1},...,x_{k,n_{k}} \right\} }[/math] then C can be written as:
[math]\displaystyle{ C=\frac{1}{n}\sum_{i=1}^{k}\sum_{j=1}^{n_{i}} \left(x_{i,j}-\frac{1}{n_{i}}X1{i}\right)\left(x_{1,i}-\frac{1}{n_{i}}X1{i}\right)^{'} }[/math]
where [math]\displaystyle{ 1_{i} }[/math] is [math]\displaystyle{ n \times 1 }[/math] vector such that:
[math]\displaystyle{ [1_{i}]_{j}= \left\{\begin{matrix} 1 & \text{patern} j \in \text{chunklet} i \\ 0 & \text{otherwise} \end{matrix}\right. }[/math]
and [math]\displaystyle{ I_{i}=diag\left(1_{i}\right) }[/math].
using the above notations C can be simplified to the form [math]\displaystyle{ C=\frac{1}{n}XHX^{'} }[/math]
where [math]\displaystyle{ H=\sum_{i=1}^{k}\left(I_{i}-\frac{1}{n_{i}}1_{i}1_{i}^{'}\right) }[/math]
for the issue of non-singularity, for small [math]\displaystyle{ \epsilon }[/math] let [math]\displaystyle{ \hat{C}=C+\epsilon I }[/math] then we can find the inverse of [math]\displaystyle{ \hat{C} }[/math] which is
[math]\displaystyle{ \hat{C}^{-1}=\frac{1}{\epsilon}I-\frac{1}{n \epsilon^{2}}XH \left(X^{'}XH \right)^{-1}X^{'} }[/math]
Therefor the the inner product of transformed [math]\displaystyle{ x }[/math] and [math]\displaystyle{ y }[/math] is
[math]\displaystyle{ \left(\hat{C}^{-\frac{1}{2}}x\right)^{'} \left(\hat{C}^{-\frac{1}{2}}x\right)= x^{'} \hat{C}^{-1} y= x^{'} \left( \frac{1}{\epsilon}I-\frac{1}{n \epsilon^{2}}XH \left(X^{'}XH \right)^{-1}X^{'} \right) y }[/math]
Now if RCA operates in feature [math]\displaystyle{ \mathcal{F} }[/math] with corresponding kernel [math]\displaystyle{ l }[/math] then the inner product between nonlinear transformations [math]\displaystyle{ \varphi (x) }[/math] and [math]\displaystyle{ \varphi (y) }[/math] after running RCA in [math]\displaystyle{ \mathcal{F} }[/math] is:
[math]\displaystyle{ \tilde{l}(x,y)=\frac{1}{\epsilon}l(x,y)-l_{x}^{'} \left( \frac{1}{n \epsilon^{2}}H \left( I+\frac{1}{n \epsilon}LH \right)^{-1} \right) l_{x} }[/math]
where [math]\displaystyle{ L=\left[ l(x_{i},x_{j}) \right]_{ij} }[/math], [math]\displaystyle{ l_{x}=\left[ l(x_{1,1},x),...,l(x_{k,n_{k}},x) \right]^{'} }[/math] and [math]\displaystyle{ l_{y}=\left[ l(x_{1,1},y),...,l(x_{k,n_{k}},y) \right]^{'} }[/math]
Experimental Results: Application to Clustering
The main goal of this method is to utilize the side information in the form of equivalence relations to improve the performance of unsupervised learning techniques. To test the proposed the above RCA algorithm and for the sake of comparison of our results by Xing et al. we used six data sets from UC Irvine repository which were used in <ref> E. Xing, A. Ng, M. Jordan, and S. Russell, "Distance metric learning with application to clustering with side-information", Advances in Neural Information Processing Systems, 2002. </ref>. Similar to what they have in their paper, we are given as set S of pairwise similarity constraints; Having this data set, we performed the following clustering algorithms:
1. K-means using the default Euclidean metric (i.e. using no side information) .
2. Constrained K-means: K-means subject to points [math]\displaystyle{ \mathbf{(x_i,x_j) \in S } }[/math] always being assigned to the same cluster (Wagstaff et al. ,2001).
3. Constrained K-means + metric proposed by (Xing et al., 2002): Constrained K-means using the distance metric proposed in (Xing et al., 2002), which is learned from S.
4. Constrained K-means + RCA: Constrained K-means using the RCA distance metric learned from S.
5. EM: Expectation Maximization of a Gaussian Mixture model (using no side-information).
6. Constrained EM: EM using side-information in the form of equivalence constraints (Hertz et al., 2002; Shental et al., 2003), when using RCA distance metric as an initial metric.
Following (Xing et al., 2002) a normalized accuracy score is used to evaluate the partitions obtained by the different clustering algorithms which we pointed out in the above six methods. More specifically, in the case of 2-cluster data the accuracy measure used can be written as:
where [math]\displaystyle{ \mathbf{1\{.\}} }[/math] is the indicator function, [math]\displaystyle{ \mathbf{\{\hat{c_i}\}_{i=1}^m} }[/math] is the cluster to which point [math]\displaystyle{ \mathbf{x_i} }[/math] is assigned by the clustering algorithm, and [math]\displaystyle{ \mathbf{c_i} }[/math] is the "correct" or desired assignment. The above score can be regarded as computing the probability that the algorithm's assignment [math]\displaystyle{ \mathbf{\hat{c}} }[/math] of two randomly drawn points [math]\displaystyle{ \mathbf{x_i} }[/math] and [math]\displaystyle{ \mathbf{x_j} }[/math] agrees with the "true" asignment [math]\displaystyle{ \mathbf{c} }[/math].
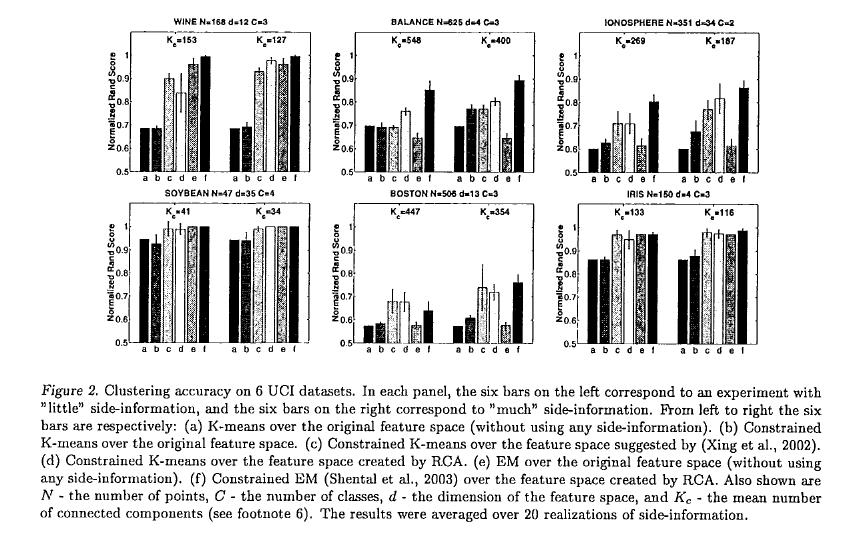
As it has been shown in this experiment, by introducing the constraints into am EM formulation, not only the negative constraints is handled, but also the RCA results has been improved dramatically in the case (f).
Similar to what (Xing et al., 2002) have done, we tested our method using two conditions:
I) Using "little" side-information [math]\displaystyle{ \mathbf{S} }[/math] II) Using "much" side-information.
In all of four experiments we sued K-means with multiple restarts. We have showed the results of all algorithms described above when we use the two conditions of "little" and "much" side-information.
As it can be seen clearly in results, using RCA as a distance measure has a significant impact on improving the results over the original K-means algorithm. Our results compared to (Xing et al., 2002) indicate that RCA achieves similar results. In this respect it should be noted that the RCA metric computation is a single step efficient computation, whereas the method presented in (Xing et al., 2002) requires gradient descent and iterative projections.
References
<references/>