Graph Structure of Neural Networks: Difference between revisions
| Line 68: | Line 68: | ||
== top architectures can be identified efficiently == | == top architectures can be identified efficiently == | ||
According to the graphs we have in the Key Results. We can see that there is a "sweet spot" and therefore, we do not have to train the entire data set for a large value of epoch. We can take a sample of the data | According to the graphs we have in the Key Results. We can see that there is a "sweet spot" and therefore, we do not have to train the entire data set for a large value of epoch or a really large sample. We can take a sample of the data around 52 graphs that would have a correlation of 0.9 which indicates that fewer samples are needed for a similar analysis in practice. Within 3 epochs, the correlation between the variables is already high enough for future computation. | ||
== well-performing neural networks have graph structure surprisingly similar to those of real biological neural networks== | == well-performing neural networks have graph structure surprisingly similar to those of real biological neural networks== | ||
Revision as of 22:32, 16 November 2020
Presented By
Xiaolan Xu, Robin Wen, Yue Weng, Beizhen Chang
Introduction
During the study of Neural Networks, it is especially important to build a relation between a neural network’s accuracy and its underlying graph structure. The natural choice is to use computational graph representation but it has many limitations such as lack of generality and disconnection with biology/neuroscience.
Thus, we develop a new way of representing a neural network as a graph, which we call a relational graph. Our key insight is to focus on message exchange, rather than just on directed data flow. As a simple example, for a fixed-width fully-connected layer, we can represent one input channel and one output channel together as a single node, and an edge in the relational graph represents the message exchange between the two nodes. Under this formulation, using the appropriate message exchange definition, we show that the relational graph can represent many types of neural network layers.
Neural Network as Relational Graph
First, we define a graph [math]\displaystyle{ G =(V; E) }[/math] by its node set [math]\displaystyle{ V =\{v_1,...,v_n\} }[/math] and edge set [math]\displaystyle{ E \subseteq \{(v_i,v_j)|v_i,v_j\in V\} }[/math]. We assume that each node $v$ correpsonds with a feature [math]\displaystyle{ x_v }[/math], which might be scalar, vector or tensor quantity. All nodes have self edges, that is [math]\displaystyle{ (v_i,v_i)\in E }[/math], and the graph we consider here is undirected.
We call graph [math]\displaystyle{ G }[/math] a relational graph, when it is associated with message exchanges between neurons. (It is not necessary for one node in neural network to correspond with one node in the relational graph.) The association can be more complicated.
First: Graph
Second: Graph with Message Exchange
Message Exchange: Message Function + Aggregation Function
Round of Message Excnage
Specifically, a message exchange is defined by a message function and an aggregation function. A meesage function takes a node's feature [math]\displaystyle{ x_v }[/math] and then outputs an measage.
An aggregation function: taking a set of messages and then output the updated node feature. Basically the aggregation function descirbes the way how the message is combined.
At each round of message exchange, each node sends messages to its neighbors and aggregates incoming messages from its neighbors. This start to look like a neural network.
Each message is transformed at each edge through a message function [math]\displaystyle{ f(·) }[/math], then they are aggregated at each node via an aggregation function [math]\displaystyle{ AGG(·) }[/math]. Suppose we conduct [math]\displaystyle{ r }[/math] rounds of message exchange, then the [math]\displaystyle{ r^{th} }[/math] round of message exchange for a node [math]\displaystyle{ v }[/math] can be described as
where [math]\displaystyle{ \mathbf{x}^{(r+1)} }[/math] is the feature of of the [math]\displaystyle{ v }[/math] node in the relational graph after the [math]\displaystyle{ r^{th} }[/math] round of update. [math]\displaystyle{ u,v }[/math]
Parameter Definition
(1) Clustering Coefficient
(2) Average Path Length
Experimental Setup (Section 4 in the paper)
Discussions and Conclusions
Section 5 of the paper summarize the result of experiment among multiple different relational graphs through sampling and analyzing.
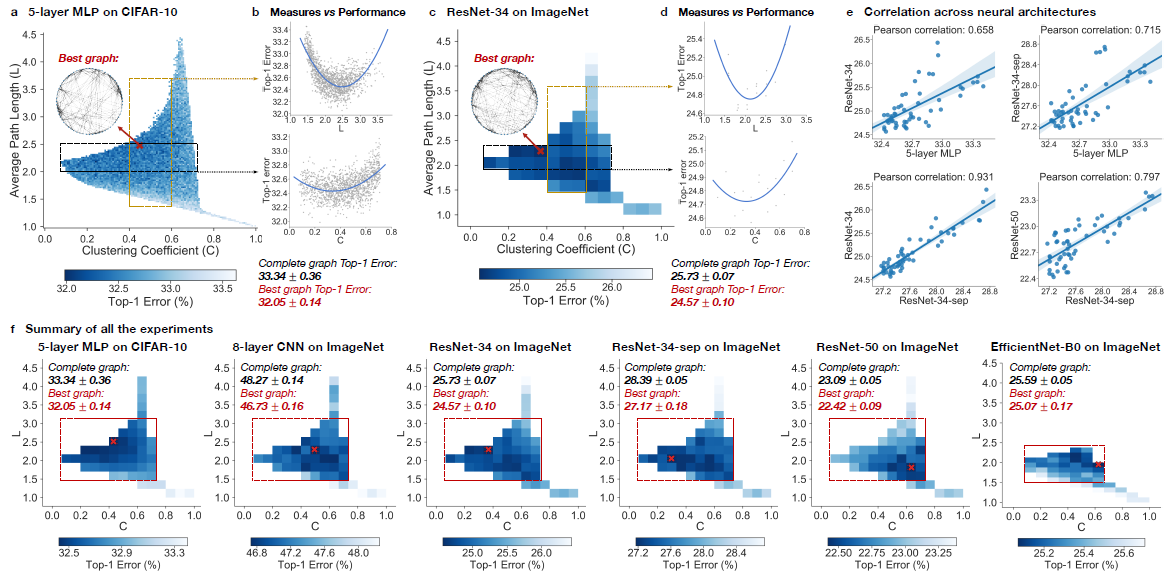
Neural networks performance depends on its structure
In the experiment, top-1 errors are going to be used to measure the performance of the model. The parameters of the models are average path length and clustering coefficient. Heat maps was created to illustrate the difference of predictive performance among possible average path length and clustering coefficient. In Figure ???, The darker area represents a smaller top-1 error which indicate the model perform better than other area. Compare with the complete graph which has A = 1 and C = 1, The best performing relational graph can outperform the complete graph baseline by 1.4% top-1 error for MLP on CIFAR-10, and 0.5% to 1.2% for models on ImageNet. Hence it is an indicator that the predictive performance of neural network highly depends on the graph structure, or equivalently that completed graph does not always preform the best.
Sweet spot where performance is significantly improved
To reduce the training noise, the 3942 graphs that in the sample had been grouped into 52 bin, each bin had been colored based on the average performance of graphs that fall into the bin. Based on the heat map, the well-performing graphs tend to cluster into a special spot that the paper called “sweet spot” shown in the red rectangle.
Relationship between neural network’s performance and parameters
When we visualize the heat map, we can see that there are no significant jump of performance that occurred as small change of clustering coefficient and average path length. If one of the variable is fixed in a small range, it is observed that a second degree polynomial is a good visualization tools for the overall trend. Therefore, both clustering coefficient and average path length are highly related with neural network performance by a U-shape.
Consistency among many different tasks and datasets
It is observed that the results are consistent through different point of view. Among multiple architecture dataset, it is observed that the clustering coefficient within [0.1,0.7] and average path length with in [1.5,3] consistently outperform the baseline complete graph.
Among different dataset with network that has similar clustering coefficient and average path length, the results are correlated, The paper mentioned that ResNet-34 is much more complex than 5-layer MLP but a fixed set relational graphs would perform similarly in both setting, with Pearson correlation of 0.658, the p-value for the Null hypothesis is less than 10^-8.
top architectures can be identified efficiently
According to the graphs we have in the Key Results. We can see that there is a "sweet spot" and therefore, we do not have to train the entire data set for a large value of epoch or a really large sample. We can take a sample of the data around 52 graphs that would have a correlation of 0.9 which indicates that fewer samples are needed for a similar analysis in practice. Within 3 epochs, the correlation between the variables is already high enough for future computation.
well-performing neural networks have graph structure surprisingly similar to those of real biological neural networks
The way we define relational graphs and average length in the graph is similar to the way information is exchanged in network science. The biological neural network also has a similar relational graph representation and graph measure with the best-performing relational graph.
Critique
1. The experiment is only measuring on a single data set which might impact the conclusion since this is not representative enough. 2. When we are fitting the model, training data should be randomized in each epoch to reduce the noise.