stat841f11: Difference between revisions
(Added lecture dates to sections.) |
(→LDA and QDA: - added description at beginning.) |
||
| Line 92: | Line 92: | ||
The former reflects a '''Bayesian''' approach. The Bayesian approach uses previous beliefs and observed data (e.g., the random variable <math>\ X </math>) to determine the probability distribution of the parameter of interest (e.g., the random variable <math>\ Y </math>). The probability, according to Bayesians, is a ''degree of belief'' in the parameter of interest taking on a particular value (e.g., <math>\ Y=1 </math>), given a particular observation (e.g., <math>\ X=x </math>). Historically, the difficulty in this approach lies with determining the posterior distribution, however, more recent methods such as '''Markov Chain Monte Carlo (MCMC)''' allow the Bayesian approach to be implemented <ref name="PCAustin">P. C. Austin, C. D. Naylor, and J. V. Tu, "A comparison of a Bayesian vs. a frequentist method for profiling hospital performance," ''Journal of Evaluation in Clinical Practice'', 2001</ref>. | The former reflects a '''Bayesian''' approach. The Bayesian approach uses previous beliefs and observed data (e.g., the random variable <math>\ X </math>) to determine the probability distribution of the parameter of interest (e.g., the random variable <math>\ Y </math>). The probability, according to Bayesians, is a ''degree of belief'' in the parameter of interest taking on a particular value (e.g., <math>\ Y=1 </math>), given a particular observation (e.g., <math>\ X=x </math>). Historically, the difficulty in this approach lies with determining the posterior distribution, however, more recent methods such as '''Markov Chain Monte Carlo (MCMC)''' allow the Bayesian approach to be implemented <ref name="PCAustin">P. C. Austin, C. D. Naylor, and J. V. Tu, "A comparison of a Bayesian vs. a frequentist method for profiling hospital performance," ''Journal of Evaluation in Clinical Practice'', 2001</ref>. | ||
The latter reflects a '''Frequentist''' approach. The Frequentist approach assumes that the probability distribution, including the mean, variance, etc., is fixed for the parameter of interest (e.g., the variable <math>\ Y </math>, which is ''not'' random). The observed data (e.g., the random variable <math>\ X </math>) is simply a ''sampling'' of a far larger population of possible observations. Thus, a certain repeatability or ''frequency'' is expected in the observed data. If it were possible to make an infinite number of observations, then the true probability distribution of the parameter of interest can be found. In general, frequentists use a technique called '''hypothesis testing''' to compare a ''null hypothesis'' (e.g. an assumption that the mean of the probability distribution is <math>\ \mu_0 </math>) to an alternative hypothesis (e.g. assuming that the mean of the probability distribution is larger than <math>\ \mu_0 </math>) <ref name="PCAustin"/>. For more information on hypothesis testing see <ref>R. Levy, "Frequency hypothesis testing, and contingency tables" class notes for LING251, Department of Linguistics, University of California, 2007 [http://idiom.ucsd.edu/~rlevy/lign251/fall2007/lecture_8.pdf] </ref>. | The latter reflects a '''Frequentist''' approach. The Frequentist approach assumes that the probability distribution, including the mean, variance, etc., is fixed for the parameter of interest (e.g., the variable <math>\ Y </math>, which is ''not'' random). The observed data (e.g., the random variable <math>\ X </math>) is simply a ''sampling'' of a far larger population of possible observations. Thus, a certain repeatability or ''frequency'' is expected in the observed data. If it were possible to make an infinite number of observations, then the true probability distribution of the parameter of interest can be found. In general, frequentists use a technique called '''hypothesis testing''' to compare a ''null hypothesis'' (e.g. an assumption that the mean of the probability distribution is <math>\ \mu_0 </math>) to an alternative hypothesis (e.g. assuming that the mean of the probability distribution is larger than <math>\ \mu_0 </math>) <ref name="PCAustin"/>. For more information on hypothesis testing see <ref>R. Levy, "Frequency hypothesis testing, and contingency tables" class notes for LING251, Department of Linguistics, University of California, 2007. Available: [http://idiom.ucsd.edu/~rlevy/lign251/fall2007/lecture_8.pdf http://idiom.ucsd.edu/~rlevy/lign251/fall2007/lecture_8.pdf] </ref>. | ||
There was some class discussion on which approach should be used. Both the ease of computation and the validity of both approaches were discussed. A main point that was brought up in class is that Frequentists consider X to be a random variable, but they do not consider Y to be a random variable because it has to take on one of the values from a fixed set (in the above case it would be either 0 or 1 and there is only one ''correct'' label for a given value X=x). Thus, from a Frequentist's perspective it does not make sense to talk about the probability of Y. This is actually a grey area and sometimes ''Bayesians'' and ''Frequentists'' use each others' approaches. So using ''Bayes' rule'' doesn't necessarily mean you're a ''Bayesian''. Overall, the question remains unresolved. | There was some class discussion on which approach should be used. Both the ease of computation and the validity of both approaches were discussed. A main point that was brought up in class is that Frequentists consider X to be a random variable, but they do not consider Y to be a random variable because it has to take on one of the values from a fixed set (in the above case it would be either 0 or 1 and there is only one ''correct'' label for a given value X=x). Thus, from a Frequentist's perspective it does not make sense to talk about the probability of Y. This is actually a grey area and sometimes ''Bayesians'' and ''Frequentists'' use each others' approaches. So using ''Bayes' rule'' doesn't necessarily mean you're a ''Bayesian''. Overall, the question remains unresolved. | ||
| Line 179: | Line 179: | ||
== LDA and QDA == | == LDA and QDA == | ||
'''Discriminant function analysis''' finds features that best allow discrimination between two or more classes. The approach is similar to '''analysis of Variance (ANOVA)''' in that discriminant function analysis looks at the mean values to determine if two or more classes are very different and should be separated. Once the discriminant functions (that separate two or more classes) have been determined, new data points can be classified (i.e. placed in one of the classes) based on the discriminant functions <ref> StatSoft, Inc. (2011). ''Electronic Statistics Textbook.'' [Online]. Available: [http://www.statsoft.com/textbook/discriminant-function-analysis/ http://www.statsoft.com/textbook/discriminant-function-analysis/.] </ref>. '''Linear discriminant analysis (LDA)''' and '''Quadratic discriminant analysis (QDA)''' are methods of discriminant analysis that are best applied to linearly and quadradically separable classes, respectively. '''Fisher discriminant analysis (FDA)''' another method of discriminant analysis that is different from linear discriminant analysis, but oftentimes both terms are used interchangeably. | |||
LDA ( | === LDA === | ||
The simplest method is to use approach 3 (above) and assume a parametric model for densities. Assume class conditional is Gaussian. | |||
<math>\mathcal{Y} = \{ 0,1 \}</math> assumed (i.e., 2 labels) | <math>\mathcal{Y} = \{ 0,1 \}</math> assumed (i.e., 2 labels) | ||
Revision as of 09:50, 29 September 2011
Editor Sign Up
STAT 441/841 / CM 463/763 - Tuesday, 2011/09/20
Wiki Course Notes
Students will need to contribute to the wiki for 20% of their grade. Access via wikicoursenote.com Go to editor sign-up, and use your UW userid for your account name, and use your UW email.
primary (10%) Post a draft of lecture notes within 48 hours. You will need to do this 1 or 2 times, depending on class size.
secondary (10%) Make improvements to the notes for at least 60% of the lectures. More than half of your contributions should be technical rather than editorial. There will be a spreadsheet where students can indicate what they've done and when. The instructor will conduct random spot checks to ensure that students have contributed what they claim.
Classification (Lecture: Sep. 20, 2011)
Definitions
classification: Predict a discrete random variable [math]\displaystyle{ Y }[/math] (a label) by using another random variable [math]\displaystyle{ X }[/math] (new data point) picked iid from a distribution
[math]\displaystyle{ X_i = (X_{i1}, X_{i2}, ... X_{id}) \in \mathcal{X} \subset \mathbb{R}^d }[/math] ([math]\displaystyle{ d }[/math]-dimensional vector) [math]\displaystyle{ Y_i }[/math] in some finite set [math]\displaystyle{ \mathcal{Y} }[/math]
classification rule:
[math]\displaystyle{ h : \mathcal{X} \rightarrow \mathcal{Y} }[/math]
Take new observation [math]\displaystyle{ X }[/math] and use a classification function [math]\displaystyle{ h(x) }[/math] to generate a label [math]\displaystyle{ Y }[/math]. In other words, If we fit the function [math]\displaystyle{ h(x) }[/math] with a random variable [math]\displaystyle{ X }[/math], it generates the label [math]\displaystyle{ Y }[/math] which is the class to which we predict [math]\displaystyle{ X }[/math] belongs.
Example: Let [math]\displaystyle{ \mathcal{X} }[/math] be a set of 2D images and [math]\displaystyle{ \mathcal{Y} }[/math] be a finite set of people. We want to learn a classification rule [math]\displaystyle{ h:\mathcal{X}\rightarrow\mathcal{Y} }[/math] that with small true error predicts the person who appears in the image.
true error rate for classifier [math]\displaystyle{ h }[/math] is the error with respect to the underlying distribution (that we do not know).
[math]\displaystyle{ L(h) = P(h(X) \neq Y ) }[/math]
empirical error rate (or training error rate) is the amount of error that our classification function [math]\displaystyle{ h(x) }[/math] makes on the training data.
[math]\displaystyle{ \hat{L}_n(h) = (1/n) \sum_{i=1}^{n} \mathbf{I}(h(X_i) \neq Y_i) }[/math]
where [math]\displaystyle{ \mathbf{I}() }[/math] is an indicator function. Indicator function is defined by
[math]\displaystyle{ \mathbf{I}(x) = \begin{cases} 1 & \text{if } x \text{ is true} \\ 0 & \text{if } x \text{ is false} \end{cases} }[/math]
So in this case [math]\displaystyle{ \mathbf{I}(h(X_i)\neq Y_i) = 1 }[/math] when [math]\displaystyle{ h(X_i)\neq Y_i }[/math] i.e. when misclassifications happens.
e.g., 100 new data points with known (true) labels
[math]\displaystyle{ y_1 = h(x_1) }[/math]
...
[math]\displaystyle{ y_{100} = h(x_{100}) }[/math]
To calculate the empirical error we count how many labels our function [math]\displaystyle{ h(x) }[/math] assigned incorrectly and divide by n=100
Bayes Classifier
The principle of Bayes Classifier is to calculate the posterior probability of a given object from its prior probability via Bayes formula, and then place the object in the class with the largest posterior probability<ref> http://www.wikicoursenote.com/wiki/Stat841#Bayes_Classifier </ref>.
First recall Bayes' Rule, in the format [math]\displaystyle{ P(Y|X) = \frac{P(X|Y) P(Y)} {P(X)} }[/math]
P(Y|X) : posterior , probability of [math]\displaystyle{ Y }[/math] given [math]\displaystyle{ X }[/math]
P(X|Y) : likelihood, probability of [math]\displaystyle{ X }[/math] being generated by [math]\displaystyle{ Y }[/math]
P(Y) : prior, probability of [math]\displaystyle{ Y }[/math] being selected
P(X) : marginal, probability of obtaining [math]\displaystyle{ X }[/math]
We will start with the simplest case: [math]\displaystyle{ \mathcal{Y} = \{0,1\} }[/math]
[math]\displaystyle{ r(x) = P(Y=1|X=x) = \frac{P(X=x|Y=1) P(Y=1)} {P(X=x)} = \frac{P(X=x|Y=1) P(Y=1)} {P(X=x|Y=1) P(Y=1) + P(X=x|Y=0) P(Y=0)} }[/math]
Bayes' rule can be approached by computing either:
1) The posterior: [math]\displaystyle{ \ P(Y=1|X=x) }[/math] and [math]\displaystyle{ \ P(Y=0|X=x) }[/math] or
2) The likelihood: [math]\displaystyle{ \ P(X=x|Y=1) }[/math] and [math]\displaystyle{ \ P(X=x|Y=0) }[/math]
The former reflects a Bayesian approach. The Bayesian approach uses previous beliefs and observed data (e.g., the random variable [math]\displaystyle{ \ X }[/math]) to determine the probability distribution of the parameter of interest (e.g., the random variable [math]\displaystyle{ \ Y }[/math]). The probability, according to Bayesians, is a degree of belief in the parameter of interest taking on a particular value (e.g., [math]\displaystyle{ \ Y=1 }[/math]), given a particular observation (e.g., [math]\displaystyle{ \ X=x }[/math]). Historically, the difficulty in this approach lies with determining the posterior distribution, however, more recent methods such as Markov Chain Monte Carlo (MCMC) allow the Bayesian approach to be implemented <ref name="PCAustin">P. C. Austin, C. D. Naylor, and J. V. Tu, "A comparison of a Bayesian vs. a frequentist method for profiling hospital performance," Journal of Evaluation in Clinical Practice, 2001</ref>.
The latter reflects a Frequentist approach. The Frequentist approach assumes that the probability distribution, including the mean, variance, etc., is fixed for the parameter of interest (e.g., the variable [math]\displaystyle{ \ Y }[/math], which is not random). The observed data (e.g., the random variable [math]\displaystyle{ \ X }[/math]) is simply a sampling of a far larger population of possible observations. Thus, a certain repeatability or frequency is expected in the observed data. If it were possible to make an infinite number of observations, then the true probability distribution of the parameter of interest can be found. In general, frequentists use a technique called hypothesis testing to compare a null hypothesis (e.g. an assumption that the mean of the probability distribution is [math]\displaystyle{ \ \mu_0 }[/math]) to an alternative hypothesis (e.g. assuming that the mean of the probability distribution is larger than [math]\displaystyle{ \ \mu_0 }[/math]) <ref name="PCAustin"/>. For more information on hypothesis testing see <ref>R. Levy, "Frequency hypothesis testing, and contingency tables" class notes for LING251, Department of Linguistics, University of California, 2007. Available: http://idiom.ucsd.edu/~rlevy/lign251/fall2007/lecture_8.pdf </ref>.
There was some class discussion on which approach should be used. Both the ease of computation and the validity of both approaches were discussed. A main point that was brought up in class is that Frequentists consider X to be a random variable, but they do not consider Y to be a random variable because it has to take on one of the values from a fixed set (in the above case it would be either 0 or 1 and there is only one correct label for a given value X=x). Thus, from a Frequentist's perspective it does not make sense to talk about the probability of Y. This is actually a grey area and sometimes Bayesians and Frequentists use each others' approaches. So using Bayes' rule doesn't necessarily mean you're a Bayesian. Overall, the question remains unresolved.
The Bayes Classifier uses [math]\displaystyle{ \ P(Y=1|X=x) }[/math]
[math]\displaystyle{ P(Y=1|X=x) = \frac{P(X=x|Y=1) P(Y=1)} {P(X=x|Y=1) P(Y=1) + P(X=x|Y=0) P(Y=0)} }[/math]
P(Y=1) : the prior, based on belief/evidence beforehand
denominator : marginalized by summation
[math]\displaystyle{ h(x) = \begin{cases} 1 \ \ \hat{r}(x) \gt 1/2 \\ 0 \ \ otherwise \end{cases} }[/math]
The set [math]\displaystyle{ \mathcal{D}(h) = \{ x : P(Y=1|X=x) = P(Y=0|X=x)... \} }[/math]
which defines a decision boundary.
Theorem: Bayes rule is optimal. I.e., if h is any other classification rule,
then [math]\displaystyle{ L(h^*) \lt = L(h) }[/math]
(This is to be proved in homework.)
Why then do we need other classfication methods? A: Because X densities are often/typically unknown. I.e., [math]\displaystyle{ f_k(x) }[/math] and/or [math]\displaystyle{ \pi_k }[/math] unknown.
[math]\displaystyle{ P(Y=k|X=x) = \frac{P(X=x|Y=k)P(Y=k)} {P(X=x)} = \frac{f_k(x) \pi_k} {\sum_k f_k(x) \pi_k} }[/math] f_k(x) is referred to as the class conditional distribution (~likelihood).
Therefore, we rely on some data to estimate quantities.
Three Main Approaches
1. Empirical Risk Minimization: Choose a set of classifiers H (e.g., line, neural network) and find [math]\displaystyle{ h^* \in H }[/math] that minimizes (some estimate of) L(h).
2. Regression: Find an estimate ([math]\displaystyle{ \hat{r} }[/math]) of function [math]\displaystyle{ r }[/math] and define [math]\displaystyle{ h(x) = \begin{cases} 1 \ \ \hat{r}(x) \gt 1/2 \\ 0 \ \ otherwise \end{cases} }[/math]
The [math]\displaystyle{ 1/2 }[/math] in the expression above is a threshold set for the regression prediction output.
In general regression refers to finding a continuous, real valued y. The problem here is more difficult, because of the restricted domain (y is a set of discrete label values).
3. Density Estimation: Estimate [math]\displaystyle{ P(X=x|Y=0) }[/math] from [math]\displaystyle{ X_i }[/math]'s for which [math]\displaystyle{ Y_i = 0 }[/math] Estimate [math]\displaystyle{ P(X=x|Y=1) }[/math] from [math]\displaystyle{ X_i }[/math]'s for which [math]\displaystyle{ Y_i = 1 }[/math] and let [math]\displaystyle{ P(Y=?) = (1/n) \sum_{i=1}^{n} Y_i }[/math]
Define [math]\displaystyle{ \hat{r}(x) = \hat{P}(Y=1|X=x) }[/math] and [math]\displaystyle{ h(x) = \begin{cases} 1 \ \ \hat{r}(x) \gt 1/2 \\ 0 \ \ otherwise \end{cases} }[/math]
It is possible that there may not be enough data to estimate from for density estimation. But the main problem lies with high dimensional spaces, as the estimation results may not be good (high error rate) and sometimes even infeasible. The term curse of dimensionality was coined by Bellman <ref>R. E. Bellman, Dynamic Programming. Princeton University Press, 1957</ref> to describe this problem.
As the dimension of the space goes up, the learning requirements go up exponentially.
To Learn more about methods for handling high-dimensional data <ref> https://docs.google.com/viewer?url=http%3A%2F%2Fwww.bios.unc.edu%2F~dzeng%2FBIOS740%2Flecture_notes.pdf</ref>
Multi-Class Classification
Generalize to case Y takes on k>2 values.
Theorem: [math]\displaystyle{ Y \in \mathcal{Y} = {1,2,..., k} }[/math] optimal rule
[math]\displaystyle{ h*(x) = argmax_k P }[/math]
where [math]\displaystyle{ P(Y=k|X=x) = \frac{f_k(x) \pi_k} {\sum_r f_r \pi_r} }[/math]
LDA and QDA
Discriminant function analysis finds features that best allow discrimination between two or more classes. The approach is similar to analysis of Variance (ANOVA) in that discriminant function analysis looks at the mean values to determine if two or more classes are very different and should be separated. Once the discriminant functions (that separate two or more classes) have been determined, new data points can be classified (i.e. placed in one of the classes) based on the discriminant functions <ref> StatSoft, Inc. (2011). Electronic Statistics Textbook. [Online]. Available: http://www.statsoft.com/textbook/discriminant-function-analysis/. </ref>. Linear discriminant analysis (LDA) and Quadratic discriminant analysis (QDA) are methods of discriminant analysis that are best applied to linearly and quadradically separable classes, respectively. Fisher discriminant analysis (FDA) another method of discriminant analysis that is different from linear discriminant analysis, but oftentimes both terms are used interchangeably.
LDA
The simplest method is to use approach 3 (above) and assume a parametric model for densities. Assume class conditional is Gaussian.
[math]\displaystyle{ \mathcal{Y} = \{ 0,1 \} }[/math] assumed (i.e., 2 labels)
[math]\displaystyle{ h(x) = \begin{cases} 1 \ \ P(Y=1|X=x) \gt P(Y=0|X=x) \\ 0 \ \ otherwise \end{cases} }[/math]
[math]\displaystyle{ P(Y=1|X=x) = \frac{f_1(x) \pi_1} {\sum_k f_k \pi_k} \ \ }[/math] (denom = P(x))
1) Assume Gaussian distributions
[math]\displaystyle{ f_k(x) = \frac{1}{(2\pi)^{-d/2} |\Sigma_k|^{-1/2}} exp(-(1/2)(\mathbf{x} - \mathbf{\mu_k}) \Sigma_k^{-1}(\mathbf{x}-\mathbf{\mu_k}) ) }[/math]
must compare [math]\displaystyle{ \frac{f_1(x) \pi_1} {p(x)} }[/math] with [math]\displaystyle{ \frac{f_0(x) \pi_0} {p(x)} }[/math] Note that the p(x) denom can be ignored: [math]\displaystyle{ f_1(x) \pi_1 }[/math] with [math]\displaystyle{ f_0(x) \pi_0 }[/math]
To find the decision boundary, set [math]\displaystyle{ f_1(x) \pi_1 = f_0(x) \pi_0 }[/math]
2) Assume [math]\displaystyle{ \Sigma_1 = \Sigma_0 }[/math], we can use [math]\displaystyle{ \Sigma = \Sigma_0 = \Sigma_1 }[/math].
Cancel [math]\displaystyle{ (2\pi)^{-d/2} |\Sigma_k|^{-1/2} }[/math] from both sides.
Take log of both sides.
Subtract one side from both sides, leaving zero on one side.
[math]\displaystyle{ -(1/2)(\mathbf{x} - \mathbf{\mu_1})^T \Sigma^{-1} (\mathbf{x}-\mathbf{\mu_1}) + log(\pi_1) - [-(1/2)(\mathbf{x} - \mathbf{\mu_0})^T \Sigma^{-1} (\mathbf{x}-\mathbf{\mu_0}) + log(\pi_0)] = 0 }[/math]
[math]\displaystyle{ (1/2)[-\mathbf{x}^T \Sigma^{-1}\mathbf{x} - \mathbf{\mu_1}^T \Sigma^{-1} \mathbf{\mu_1} + 2\mathbf{\mu_1}^T \Sigma^{-1} \mathbf{x}
+ \mathbf{x}^T \Sigma^{-1}\mathbf{x} + \mathbf{\mu_0}^T \Sigma^{-1} \mathbf{\mu_0} - 2\mathbf{\mu_0}^T \Sigma^{-1} \mathbf{x} ]
+ log(\pi_1/\pi_0) = 0 }[/math]
Cancelling out the terms quadratic in [math]\displaystyle{ \mathbf{x} }[/math] and rearranging results in
[math]\displaystyle{ (1/2)[-\mathbf{\mu_1}^T \Sigma^{-1} \mathbf{\mu_1} + \mathbf{\mu_0}^T \Sigma^{-1} \mathbf{\mu_0} + (2\mathbf{\mu_1}^T \Sigma^{-1} - 2\mathbf{\mu_0}^T \Sigma^{-1}) \mathbf{x}] + log(\pi_1/\pi_0) = 0 }[/math]
We can see that the first pair of terms is constant, and the second pair is linear in x.
So that we end up with something of the form
[math]\displaystyle{ ax + b = 0 }[/math].
LDA and QDA Continued (Lecture: Sep. 22, 2011)
If we relax assumption 2 (eg. [math]\displaystyle{ \Sigma_1 != \Sigma_0 }[/math]) then we get a quadratic equation that can be written as [math]\displaystyle{ {x}^Ta{x}+b{x} + c = 0 }[/math]
Generalizing LDA and QDA
Theorem:
Suppose that [math]\displaystyle{ \,Y \in \{1,\dots,K\} }[/math], if [math]\displaystyle{ \,f_k(x) = Pr(X=x|Y=k) }[/math] is Gaussian, the Bayes Classifier rule is
- [math]\displaystyle{ \,h^*(x) = \arg\max_{k} \delta_k(x) }[/math]
Where
[math]\displaystyle{ \,\delta_k(x) = - \frac{1}{2}log(|\Sigma_k|) - \frac{1}{2}(x-\mu_k)^\top\Sigma_k^{-1}(x-\mu_k) + log (\pi_k) }[/math]
When the Gaussian variances are equal [math]\displaystyle{ \Sigma_1 = \Sigma_0 }[/math] (e.g. LDA), then
[math]\displaystyle{ \,\delta_k(x) = x^\top\Sigma^{-1}\mu_k - \frac{1}{2}\mu_k^\top\Sigma^{-1}\mu_k + log (\pi_k) }[/math]
In practice
We estimate the prior to be the chance that a random item from the collection belongs to set k, e.g.
[math]\displaystyle{ \,\hat{\pi_k} = \hat{Pr}(y=k) = \frac{n_k}{n} }[/math]
The mean to be the average item in set k, e.g.
[math]\displaystyle{ \,\hat{\mu_k} = \frac{1}{n_k}\sum_{i:y_i=k}x_i }[/math]
and calculate the covariance of each class e.g.
[math]\displaystyle{ \,\hat{\Sigma_k} = \frac{1}{n_k}\sum_{i:y_i=k}(x_i-\hat{\mu_k})(x_i-\hat{\mu_k})^\top }[/math]
If we wish to use LDA we must calculate a common covariance, so we average all the covariances e.g.
[math]\displaystyle{ \,\Sigma=\frac{\sum_{r=1}^{k}(n_r\Sigma_r)}{n} }[/math]
Where: [math]\displaystyle{ \,n_r }[/math] is the number of data points in class [math]\displaystyle{ \,r }[/math], [math]\displaystyle{ \,\Sigma_r }[/math] is the covariance of class [math]\displaystyle{ \,r }[/math], [math]\displaystyle{ \,n }[/math] is the total number of data points, and [math]\displaystyle{ \,k }[/math] is the number of classes.
Computation
For QDA we need to calculate: [math]\displaystyle{ \,\delta_k(x) = - \frac{1}{2}log(|\Sigma_k|) - \frac{1}{2}(x-\mu_k)^\top\Sigma_k^{-1}(x-\mu_k) + log (\pi_k) }[/math]
Lets first consider when [math]\displaystyle{ \, \Sigma_k = I, \forall k }[/math]. This is the case where each distribution is spherical, around the mean point.
Case 1
When [math]\displaystyle{ \, \Sigma_k = I }[/math]
We have:
[math]\displaystyle{ \,\delta_k = - \frac{1}{2}log(|I|) - \frac{1}{2}(x-\mu_k)^\top I(x-\mu_k) + log (\pi_k) }[/math]
but [math]\displaystyle{ \ \log(|I|)=\log(1)=0 }[/math]
and [math]\displaystyle{ \, (x-\mu_k)^\top I(x-\mu_k) = (x-\mu_k)^\top(x-\mu_k) }[/math] is the squared Euclidean distance between two points [math]\displaystyle{ \,x }[/math] and [math]\displaystyle{ \,\mu_k }[/math]
Thus in this condition, a new point can be classified by its distance away from the center of a class, adjusted by some prior.
Further, for two-class problem with equal prior, the discriminating function would be the bisector of the 2-class's means.
Case 2
When [math]\displaystyle{ \, \Sigma_k \neq I }[/math]
Using the Singular Value Decomposition(SVD) of [math]\displaystyle{ \, \Sigma_k }[/math] we get [math]\displaystyle{ \, \Sigma_k = U_kS_kV_k^\top }[/math]
but since [math]\displaystyle{ \, \Sigma }[/math] is symmetric<ref> http://en.wikipedia.org/wiki/Covariance_matrix#Properties </ref> [math]\displaystyle{ \, \Sigma_k = U_kS_kU_k^\top }[/math]
For [math]\displaystyle{ \,\delta_k }[/math], the second term becomes what is also known as the Mahalanobis distance <ref>P. C. Mahalanobis, "On The Generalised Distance in Statistics," Proceedings of the National Institute of Sciences of India, 1936</ref> :
- [math]\displaystyle{ \begin{align} (x-\mu_k)^\top\Sigma_k^{-1}(x-\mu_k)&= (x-\mu_k)^\top U_kS_k^{-1}U_k^T(x-\mu_k)\\ & = (U_k^\top x-U_k^\top\mu_k)^\top S_k^{-1}(U_k^\top x-U_k^\top \mu_k)\\ & = (U_k^\top x-U_k^\top\mu_k)^\top S_k^{-\frac{1}{2}}S_k^{-\frac{1}{2}}(U_k^\top x-U_k^\top\mu_k) \\ & = (S_k^{-\frac{1}{2}}U_k^\top x-S_k^{-\frac{1}{2}}U_k^\top\mu_k)^\top I(S_k^{-\frac{1}{2}}U_k^\top x-S_k^{-\frac{1}{2}}U_k^\top \mu_k) \\ & = (S_k^{-\frac{1}{2}}U_k^\top x-S_k^{-\frac{1}{2}}U_k^\top\mu_k)^\top(S_k^{-\frac{1}{2}}U_k^\top x-S_k^{-\frac{1}{2}}U_k^\top \mu_k) \\ \end{align} }[/math]
If we think of [math]\displaystyle{ \, S_k^{-\frac{1}{2}}U_k^\top }[/math] as a linear transformation that takes points in class [math]\displaystyle{ \,k }[/math] and distributes them spherically around a point, like in case 1. Thus when we are given a new point, we can apply the modified [math]\displaystyle{ \,\delta_k }[/math] values to calculate [math]\displaystyle{ \ h^*(\,x) }[/math]. After applying the singular value decomposition, [math]\displaystyle{ \,\Sigma_k^{-1} }[/math] is considered to be an identity matrix such that
[math]\displaystyle{ \,\delta_k = - \frac{1}{2}log(|I|) - \frac{1}{2}[(S_k^{-\frac{1}{2}}U_k^\top x-S_k^{-\frac{1}{2}}U_k^\top\mu_k)^\top(S_k^{-\frac{1}{2}}U_k^\top x-S_k^{-\frac{1}{2}}U_k^\top \mu_k)] + log (\pi_k) }[/math]
and,
[math]\displaystyle{ \ \log(|I|)=\log(1)=0 }[/math]
For applying the above method with classes that have different covariance matrices (for example the covariance matrices [math]\displaystyle{ \ \Sigma_0 }[/math] and [math]\displaystyle{ \ \Sigma_1 }[/math] for the two class case), each of the covariance matrices has to be decomposed using SVD to find the according transformation. Then, each new data point has to be transformed using each transformation to compare its distance to the mean of each class (for example for the two class case, the new data point would have to be transformed by the class 1 transformation and then compared to [math]\displaystyle{ \ \mu_0 }[/math] and the new data point would also have to be transformed by the class 2 transformation and then compared to [math]\displaystyle{ \ \mu_1 }[/math]).
Principal Component Analysis (Lecture: Sep. 27, 2011)
Principal Component Analysis (PCA) is a transformation that transforms the data from a D dimensional space into a new coordinate system of dimension d, where d < D. The goal is to preserve the most of the variability in the data when switching the coordinate systems.
The new variables that form a new coordinate system are called principal components (PCs). PCs are denote by [math]\displaystyle{ u_1, u_2, ... , u_D }[/math]. Since PCs are orthogonal linear transformations of the original variables there is at most D PCs. In hope, we will only need d PCs, [math]\displaystyle{ u_1, u_2, ... , u_d }[/math], to approximate the space spanned by our values [math]\displaystyle{ x_1, x_2, ... , x_D }[/math].
Let [math]\displaystyle{ PC_j }[/math] be a linear combination of [math]\displaystyle{ x_1, x_2, ... , x_D }[/math] defined by the coefficients
[math]\displaystyle{ W^{(j)} }[/math] = [math]\displaystyle{ ( {w_1}^{(j)}, {w_2}^{(j)},...,{w_D}^{(j)} )^T }[/math]
Thus [math]\displaystyle{ u_j = {w_1}^{(j)} x_1 + {w_2}^{(j)} x_2 + ... + {w_D}^{(j)} x_D = W^{(j)^T} X }[/math]
This is a unique set up since it sets up the PCs in order from maximum to minimum variances. The first PC, [math]\displaystyle{ u_1 }[/math] is called first principal components and it has the maximum variance, thus it accounts for the most variability in the data [math]\displaystyle{ x_1, x_2, ... , x_D }[/math]. The second PC, [math]\displaystyle{ u_2 }[/math] is called second principal components and it has the second highest variance and so on until PC, [math]\displaystyle{ u_D }[/math] which has the minimum variance.
To get the first principal component, we would like to solve the following equation
[math]\displaystyle{ max (Var(W^T X)) }[/math] and solve for W that makes it true
[math]\displaystyle{ = max (W^T S W) }[/math] where S is the covariance matrix
Note: we require the following constraint, set [math]\displaystyle{ W^T W = 1 }[/math] because if there is on constraint on the length of W than there is no upper bound. Since there is a constraint now, we are searching for the direction and not the length that maximums the variance.
Lagrange Multiplier
Before we can proceed, we must review Lagrange multipliers.
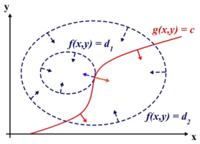
To find the maximum (or minimum) of a function [math]\displaystyle{ \displaystyle f(x,y) }[/math] subject to constraints [math]\displaystyle{ \displaystyle g(x,y) = 0 }[/math], we define a new variable [math]\displaystyle{ \displaystyle \lambda }[/math] called a Lagrange Multiplier and we form the Lagrangian,
[math]\displaystyle{ \displaystyle L(x,y,\lambda) = f(x,y) - \lambda g(x,y) }[/math]
If [math]\displaystyle{ \displaystyle (x^*,y^*) }[/math] is the max of [math]\displaystyle{ \displaystyle f(x,y) }[/math], there exists [math]\displaystyle{ \displaystyle \lambda^* }[/math] such that [math]\displaystyle{ \displaystyle (x^*,y^*,\lambda^*) }[/math] is a stationary point of [math]\displaystyle{ \displaystyle L }[/math] (partial derivatives are 0).
In addition [math]\displaystyle{ \displaystyle (x^*,y^*) }[/math] is a point in which functions [math]\displaystyle{ \displaystyle f }[/math] and [math]\displaystyle{ \displaystyle g }[/math] touch but do not cross. At this point, the tangents of [math]\displaystyle{ \displaystyle f }[/math] and [math]\displaystyle{ \displaystyle g }[/math] are parallel or gradients of [math]\displaystyle{ \displaystyle f }[/math] and [math]\displaystyle{ \displaystyle g }[/math] are parallel, such that:
[math]\displaystyle{ \displaystyle \nabla_{x,y } f = \lambda \nabla_{x,y } g }[/math]
where,
[math]\displaystyle{ \displaystyle \nabla_{x,y} f = (\frac{\partial f}{\partial x},\frac{\partial f}{\partial{y}}) \leftarrow }[/math] the gradient of [math]\displaystyle{ \, f }[/math]
[math]\displaystyle{ \displaystyle \nabla_{x,y} g = (\frac{\partial g}{\partial{x}},\frac{\partial{g}}{\partial{y}}) \leftarrow }[/math] the gradient of [math]\displaystyle{ \, g }[/math]
Example
Suppose we wish to maximize the function [math]\displaystyle{ \displaystyle f(x,y)=x-y }[/math] subject to the constraint [math]\displaystyle{ \displaystyle x^{2}+y^{2}=1 }[/math]. We can apply the Lagrange multiplier method on this example; the lagrangian is:
[math]\displaystyle{ \displaystyle L(x,y,\lambda) = x-y - \lambda (x^{2}+y^{2}-1) }[/math]
We want the partial derivatives equal to zero:
[math]\displaystyle{ \displaystyle \frac{\partial L}{\partial x}=1+2 \lambda x=0 }[/math]
[math]\displaystyle{ \displaystyle \frac{\partial L}{\partial y}=-1+2\lambda y=0 }[/math]
[math]\displaystyle{ \displaystyle \frac{\partial L}{\partial \lambda}=x^2+y^2-1 }[/math]
Solving the system we obtain 2 stationary points: [math]\displaystyle{ \displaystyle (\sqrt{2}/2,-\sqrt{2}/2) }[/math] and [math]\displaystyle{ \displaystyle (-\sqrt{2}/2,\sqrt{2}/2) }[/math]. In order to understand which one is the maximum, we just need to substitute it in [math]\displaystyle{ \displaystyle f(x,y) }[/math] and see which one as the biggest value. In this case the maximum is [math]\displaystyle{ \displaystyle (\sqrt{2}/2,-\sqrt{2}/2) }[/math].
Determining W
Use the Lagrange multiplier conversion to obtain: [math]\displaystyle{ L(W, \lambda) = W^T SW - \lambda (W^T W - 1) }[/math] where λ is a constant
Take the derivative and set it to zero:
[math]\displaystyle{ {\Delta L \over{\Delta W}} = 0 }[/math]
To obtain:
[math]\displaystyle{ 2SW - 2 \lambda W = 0 }[/math]
Rearrange to obtain:
[math]\displaystyle{ SW = \lambda W }[/math]
where W is eigenvector of S and λ is the eigenvalue of S
The [math]\displaystyle{ Var(u_1) }[/math] is maximized if [math]\displaystyle{ \lambda_1 }[/math] is the maximum eigenvalue of S and the corresponding eigenvecotor W is the first principal component. All of the PCs are generated in this way that each eigenvector of S and the corresponding eigenvalues satisfy
[math]\displaystyle{ \lambda_1 }[/math] ≥ [math]\displaystyle{ \lambda_2 }[/math] ≥ ... ≥ [math]\displaystyle{ \lambda_d }[/math] which corresponds with
[math]\displaystyle{ W_1 }[/math] ≥ [math]\displaystyle{ W_2 }[/math] ≥ ... ≥ [math]\displaystyle{ W_d }[/math]
as [math]\displaystyle{ SW= \lambda W }[/math] , and [math]\displaystyle{ W^T W=1 }[/math] , then we can write
[math]\displaystyle{ W^T SW= W^T\lambda W= \lambda W^T W =\lambda }[/math]
Meaning that W that corresponds to the maximum eigenvalue which in this case[math]\displaystyle{ \lambda_1 }[/math], will maximize the objective function or in othe words, will give the direction of the maximum variation.
so that [math]\displaystyle{ Var(u_1) }[/math] ≥ [math]\displaystyle{ Var(u_2) }[/math] ≥ ... ≥ [math]\displaystyle{ Var(u_d) }[/math]
Note that the PCs decompose the total variance in the data in the following way
[math]\displaystyle{ \sum_{i=1}^{D} Var(u_i) }[/math]
= [math]\displaystyle{ \sum_{i=1}^{D} \lambda_i) }[/math] = Tr(S) = [math]\displaystyle{ \sum_{i=1}^{D} Var(x_i) }[/math]
References
<references />
Jgpitt - 2011/09/21