probabilistic PCA with GPLVM: Difference between revisions
m (→Overview) |
|||
| Line 9: | Line 9: | ||
With dimensionality reduction, we have some high-dimensional observations, and we wish to learn a functional mapping which will transform the data from high-dimensional space into a lower-dimensional, embedded space. | With dimensionality reduction, we have some high-dimensional observations, and we wish to learn a functional mapping which will transform the data from high-dimensional space into a lower-dimensional, embedded space. | ||
With latent variable models, however, we specify a functional form <math>\,y = g(x) + \epsilon</math>, where <math>\,y</math> is a high-dimensional original data point, <math>\,x</math> is the low-dimensional mapping of <math>\,y</math>, and <math>\,\epsilon</math> is a noise model. We then estimate the parameters using [http://en.wikipedia.org/wiki/Sampling_%28statistics%29 sampling techniques] or expectation maximization (Sean Borman provides a tutorial of it [http://www.isi.edu/natural-language/teaching/cs562/2009/readings/B06.pdf here]). | With latent variable models, however, we specify a functional form <math>\,y = g(x) + \epsilon</math>, where <math>\,y</math> is a high-dimensional original data point, <math>\,x</math> is the low-dimensional mapping of <math>\,y</math>, and <math>\,\epsilon</math> is a noise model. We then estimate the parameters using [http://en.wikipedia.org/wiki/Sampling_%28statistics%29 sampling techniques] or expectation maximization (Sean Borman provides a tutorial of it [http://www.isi.edu/natural-language/teaching/cs562/2009/readings/B06.pdf here]). | ||
Similar to the deterministic case, the probabilistic formulation has a linear form when <math>g</math> is linear, and a nonlinear form when <math>g</math> is nonlinear. | Similar to the deterministic case, the probabilistic formulation has a linear form when <math>\,g</math> is linear, and a nonlinear form when <math>\,g</math> is nonlinear. | ||
Examples of probabilistic linear latent variable modelling include Probabilistic PCA, which will be discussed next, and nonlinear techniques include [http://en.wikipedia.org/wiki/Generative_topographic_map Generative Topographic Modelling] (GTM). | Examples of probabilistic linear latent variable modelling include Probabilistic PCA, which will be discussed next, and nonlinear techniques include [http://en.wikipedia.org/wiki/Generative_topographic_map Generative Topographic Modelling] (GTM). | ||
It is important to note, however, that the GPLVM does not define <math>g</math> directly; instead, <math>g</math> is defined via the kernel trick. | It is important to note, however, that the GPLVM does not define <math>g</math> directly; instead, <math>g</math> is defined via the kernel trick. | ||
Revision as of 17:43, 3 December 2010
Overview
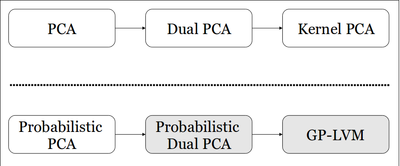
The development of principal component analysis (PCA), a statistical tool which may be used for dimensionality reduction, spans from the initial formulation of PCA 100 years ago to the more recent formulation of the dual problem (Dual PCA) and subsequent kernelization of the PCA algorithm (Kernel PCA). Inspired by this progression of the deterministic formulation of PCA, Neil Lawrence builds on a probabilistic PCA model (PPCA) developed by Tipping and Bishop, and proposes a novel dual formulation of PPCA and subsequently the Gaussian Process Latent Variable Model (GPLVM). A graphical representation of this can be see in Figure 1.
While the probabilistic approach to dimensionality reduction - latent variable model (Jeroen K. Vermunt gives details regarding this type of model here) - accomplishes the same task of dimensionality reduction, the paradigm is not entirely the same. With dimensionality reduction, we have some high-dimensional observations, and we wish to learn a functional mapping which will transform the data from high-dimensional space into a lower-dimensional, embedded space. With latent variable models, however, we specify a functional form [math]\displaystyle{ \,y = g(x) + \epsilon }[/math], where [math]\displaystyle{ \,y }[/math] is a high-dimensional original data point, [math]\displaystyle{ \,x }[/math] is the low-dimensional mapping of [math]\displaystyle{ \,y }[/math], and [math]\displaystyle{ \,\epsilon }[/math] is a noise model. We then estimate the parameters using sampling techniques or expectation maximization (Sean Borman provides a tutorial of it here). Similar to the deterministic case, the probabilistic formulation has a linear form when [math]\displaystyle{ \,g }[/math] is linear, and a nonlinear form when [math]\displaystyle{ \,g }[/math] is nonlinear. Examples of probabilistic linear latent variable modelling include Probabilistic PCA, which will be discussed next, and nonlinear techniques include Generative Topographic Modelling (GTM). It is important to note, however, that the GPLVM does not define [math]\displaystyle{ g }[/math] directly; instead, [math]\displaystyle{ g }[/math] is defined via the kernel trick.
Probabilistic PCA
In order to understand the GPLVM, we must first consider the previous work done by Tipping and Bishop in developing the PPCA. Consider a set of centred data with [math]\displaystyle{ n }[/math] observations and [math]\displaystyle{ d }[/math] dimensions, [math]\displaystyle{ \mathbf{Y} = [y_1,...,y_n]^T }[/math].
We can view this data as having a linear relationship with some embedded latent-space data [math]\displaystyle{ \mathbf{x_n} }[/math], assuming Gaussian noise, as follows:
- [math]\displaystyle{ \mathbf{y_n} = \mathbf{Wx_n} + \mathbf{n} }[/math]
where [math]\displaystyle{ x_n }[/math] is the [math]\displaystyle{ q }[/math]-dimensional latent variable associated with each observation, and [math]\displaystyle{ W \in \mathbb{R}^{d\times q} }[/math] is the transformation matrix which relates the observed space to the latent space. Assuming a Gaussian distribution for the noise with a mean of zero and a covariance of [math]\displaystyle{ \beta^{-1}\mathbf{I} }[/math], the likelihood for a particular observation [math]\displaystyle{ y_n }[/math] can be expressed as,
- [math]\displaystyle{ p(\mathbf{y_n | x_n, W},\beta) = N(\mathbf{y_n | Wx_n},\beta^{-1}\mathbf{I}) }[/math]
Here, [math]\displaystyle{ \,N }[/math] denotes the distribution function of a multivariate normal distribution for [math]\displaystyle{ y_n }[/math] with, in this case, mean [math]\displaystyle{ Wx_n }[/math] and covariance matrix [math]\displaystyle{ \beta^{-1}\mathbf{I}) }[/math].
Thus, we define a conditional probability model for the high dimensional data and, in doing so, specify a linear relationship between the latent space and data space, along with some Gaussian noise. Marginalizing both hidden variables together ([math]\displaystyle{ x_n }[/math] and [math]\displaystyle{ W }[/math]) is intractable, so we then take the standard approach of marginalizing [math]\displaystyle{ x_n }[/math], putting a Gaussian prior on [math]\displaystyle{ \mathbf{W} }[/math] and solving using maximum likelihood. Then the marginal likelihood of [math]\displaystyle{ y_n }[/math] (after marginalizing [math]\displaystyle{ x_n }[/math]) is described as,
- [math]\displaystyle{ p(\mathbf{y_n | W},\beta) = N(\mathbf{y_n |0, WW^{T}} +\beta^{-1}\mathbf{I}) }[/math]
And, assuming that the data is i.i.d., the likelihood of the full set is given as the product of the individual probabilities,
- [math]\displaystyle{ p(\mathbf{Y | W},\beta) = \prod^{N}_{n=1} p(\mathbf{y_n | W},\beta) }[/math]
We can then find [math]\displaystyle{ W }[/math] by maximizing this expression.
Tipping and Bishop do this, and find [math]\displaystyle{ W }[/math] has a closed form solution which spans the principle subspace. This is the same answer that we arrive at with PCA, where we project the data onto the subspace which maximizes the variance.
This is an acceptable formulation; however, it is only a linear model. Can it be extended to capture non-linear features? The following re-formulation as a dual and subsequent presentation of the GPLVM will show that this is in fact possible.
Dual PPCA
Dual PPCA is formulated by optimizing the latent variable [math]\displaystyle{ x_n }[/math], and marginalizing [math]\displaystyle{ \mathbf{W} }[/math]. It, in essence, follows the same steps as the work done by Bishop and Tipping. In this case, we start by defining a conjugate prior,
- [math]\displaystyle{ P(\mathbf{W}) = \prod^{D}_{d=1} N(\mathbf{W_{d,:}|0,I}) }[/math]
We then marginalize over the dimensions of [math]\displaystyle{ \mathbf{W} }[/math], rather than the observations of [math]\displaystyle{ x_n }[/math], yielding an expression for the dth column of [math]\displaystyle{ y }[/math],
- [math]\displaystyle{ p(\mathbf{y_{:,d} | X},\beta) = N(\mathbf{y_{:,d} |0, XX^{T}} +\beta^{-1}\mathbf{I}) }[/math]
This is very similar to what we saw previously, with PPCA. Here, each dimension is a multivariate normal with a covariance specified by the dot product-matrix. Assuming each dimension is independent, we can then obtain the joint likelihood of the data matrix [math]\displaystyle{ Y }[/math] as a product of each dimension. We obtain a log-likelihood objective function of the form
- [math]\displaystyle{ L = -\frac{DN}{2}\text{ln}2\pi - \frac{D}{2}\text{ln}|\mathbf{K}| - \frac{1}{2}\text{tr}(\mathbf{K^{-1}YY^T}) }[/math]
where the kernel, [math]\displaystyle{ K }[/math], is linear and of the form
- [math]\displaystyle{ \mathbf{K = XX^T} + \beta^{-1}\mathbf{I} }[/math]
Gradient descent is then applied to this objective function and a solution is obtained in the form of an eigenvalue decomposition problem [math]\displaystyle{ \mathbf{X = ULV^T} }[/math]. The solution of this likelihood function will then be the projection of the data points onto its principal subspace. Here, [math]\displaystyle{ \mathbf{U} }[/math] will be an [math]\displaystyle{ \,N \times q }[/math] matrix, with the columns being the first [math]\displaystyle{ q }[/math] eigenvectors of the matrix [math]\displaystyle{ \mathbf{YY^T} }[/math].
Note that in this formulation, each dimension of the marginal distribution can be interpreted as a Gaussian process, and each dimension of the joint marginal is a product of Gaussian processes. Thus, the objective function is expressed in terms of a linear kernel covariance.
The advantage of this dual formulation is two-fold.:
- The eigendecomposition is done on an [math]\displaystyle{ \,N \times q }[/math] matrix instead of a [math]\displaystyle{ \,d \times q }[/math] matrix, where [math]\displaystyle{ N \lt \lt d }[/math].
- [math]\displaystyle{ U }[/math] is now an inner product matrix, which can be kernelized.
Through the selection of an appropriate kernel, this technique can now be used to solve a much more broad range of dimensionality reduction problems instead of being restricted to those with linear features.
GP-LVM
GP-LVM can be accomplished by replacing the inner product kernel with one that allows for non-linear functions. We use Gaussian processes to map from a latent space, X, to the observed-data space, Y.
So, instead of a linear kernel,
- [math]\displaystyle{ k(\mathbf{x_i,x_j}) = \mathbf{x_i^Tx_j} + \beta^{-1} \delta_{i,j} }[/math]
we can consider arbitrary nonlinear kernels such as the Radial Basis Function (RBF) kernel,
- [math]\displaystyle{ k(\mathbf{x_i,x_j}) = \theta_{\mathrm{rbf}} e^{\frac{-\gamma}{2}(\mathbf{x_i - x_j)}^{T}(\mathbf{x_i - x_j)}} + \theta_{\mathrm{bias}} + \theta_{\mathrm{white}}\delta_{i,j} }[/math]
This provides a nonlinear probabilistic latent variable model - the GPLVM. However, with these nonlinear kernels, we no longer have a closed form solution.
Unifying Objective Function
We must now solve a non-convex objective function. This is is based on minimizing the Kullback-Leibler (KL) divergence between the two Gaussians which are induced by kernel matricies on the latent and data spaces:
- [math]\displaystyle{ \mathrm{KL}(N(\mathbf{z|0,S)} || N(\mathbf{z|0,K})) = \frac{1}{2} ln |\mathbf{K}| - \frac{1}{2} ln |\mathbf{S}| + \frac{1}{2} tr(\mathbf{SK^{-1}}) - \frac{N}{2} }[/math]
where [math]\displaystyle{ S }[/math] is the similarity matrix like those used in methods like MDS and Kernel PCA. This provides a unifying objective function that covers PCA, Kernel PCA, PPCA and the GPLVM.
To solve it, the author uses gradient-based optimization, and uses deterministic dimensionality reduction to generate initial values. So, not only is this nonconvex, but each evaluation of the objective function is very costly, requiring the inversion of the kernel matrix, an operation of the order [math]\displaystyle{ O(N^3) }[/math].
To speed up the optimization process, the author presents sparsification techniques to infer which subset of the data is deemed to contain the most information.
Results
The author empirically evaluates the GPLVM on several datasets. He begins with an experiment using simulated oil data, where each point is 12 dimensional, and the points can be classified into one of three flow rates---stratified, annular and homogeneous. However, the author uses only a reduced dataset of 100 points in order to make the problem computationally tractable. If we evaluate the performance of the GPLVM against PCA through classification error, using a nearest-neighbour classifier, PCA has 14 errors, while the GPLVM has four errors.
Thus, while there is some improvement in classification, it is not remarkable. This is likely due, in part, to the fact that PCA is used to initialize the GPLVM algorithm.
The author then applies the sparsified GPLVM to all 1000 oil data points; he compares it with GTM and PCA. We see that the sparsification process required to make the GPLVM tractable degrades the performance considerably.
However, while the paper shows that the GPLVM does not provide a significantly better method for classification, other domains have already put the model to use. For example, immediately after this paper was published, other researchers in the field of animation (style-based inverse kinematics) used the GPLVM on motion capture data to interpolate natural poses.
Conclusion
The GPLVM is a novel approach to probabilistic latent embedding, which is distinct from other approaches. It is probabilistic and can therefore be used to evaluate likelihoods, handle missing data and illustrate areas of confidence in the latent embedding. Further, it is nonlinear, and can consider different types of noise and kernel types. However, solving the optimization problem is difficult. It does not learn manifolds effectively, and it is computationally expensive so it does not scale well. Ultimately, however, the paper has already proved useful, particularly for missing data problems.