genetics: Difference between revisions
No edit summary |
|||
| (8 intermediate revisions by 3 users not shown) | |||
| Line 12: | Line 12: | ||
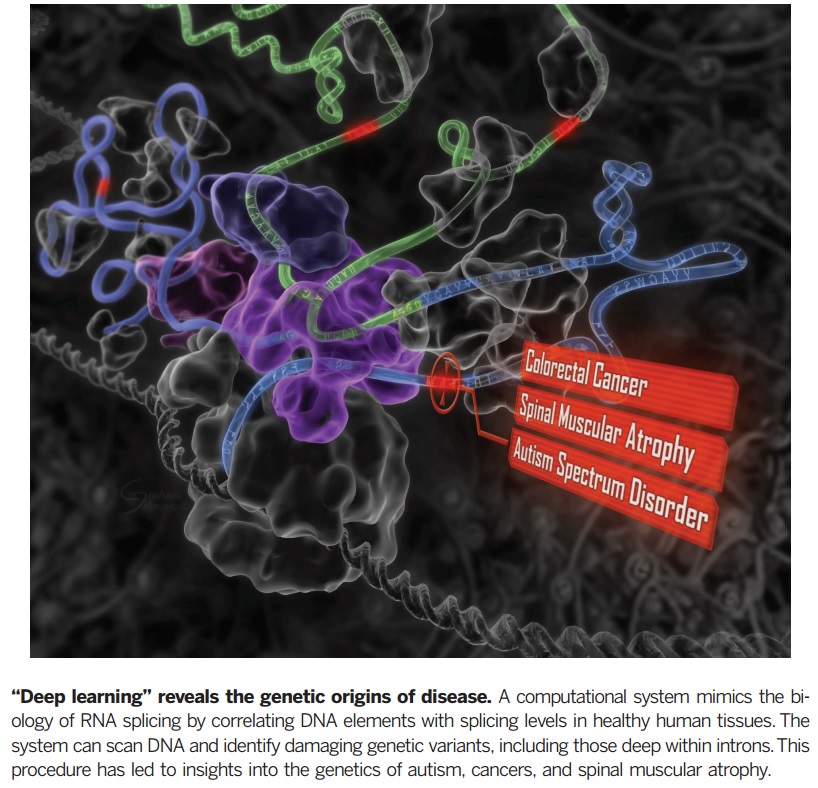
It has been a while since whole-genome sequencing been used to detect the source of disease or unwanted malignancies genetically. The idea is to find a hierarchy of mutations tending to such diseases by looking at alterations via genetic variations in the genome and particularly when they occur outside of those domains in which protein-coding happens. In the present paper, a computational method is given to detect those genetic variants which influence RNA splicing. RNA splicing is a modification of pre-messenger RNA (pre-mRNA) when introns are removed and makes the exons joined. Any type of interruptions on this important step of gene expression would lead to various kind of disease such as cancers and neurological disorders. | It has been a while since whole-genome sequencing been used to detect the source of disease or unwanted malignancies genetically. The idea is to find a hierarchy of mutations tending to such diseases by looking at alterations via genetic variations in the genome and particularly when they occur outside of those domains in which protein-coding happens. In the present paper, a computational method is given to detect those genetic variants which influence RNA splicing. RNA splicing is a modification of pre-messenger RNA (pre-mRNA) when introns are removed and makes the exons joined. Any type of interruptions on this important step of gene expression would lead to various kind of disease such as cancers and neurological disorders. | ||
[[File:Stat1.jpg]] | |||
''' | |||
== Rationale == | == Rationale == | ||
''' | ''' | ||
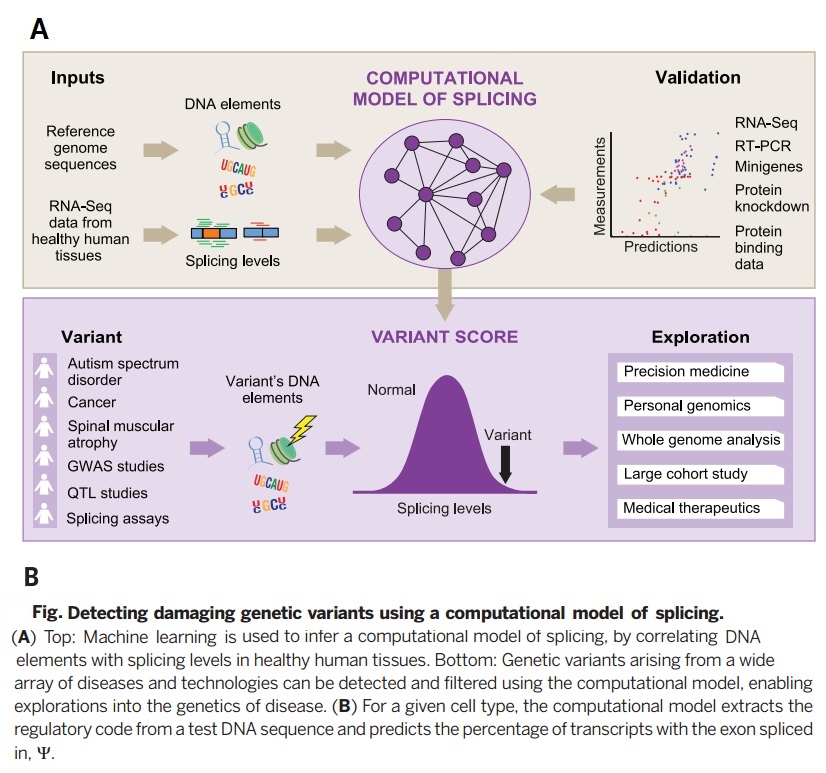
Deep learning algorithm is used to construct a computational model in which DNA sequences are inputs to predict splicing in human textures. In this model, test variants up to 300 nucleotides into an intron, can then be used to derive a score for variant alterations for splicing. | Deep learning algorithm is used to construct a computational model in which DNA sequences are inputs to predict splicing in human textures. In this model, test variants up to 300 nucleotides into an intron, can then be used to derive a score for variant alterations for splicing. The model is not biased by existing disease annotations or population data and was derived in such a way that it can be used to study diverse diseases and disorders and to determine the consequences of common, rare, and even spontaneous variants. | ||
[[File:Stat3.jpg]] | |||
''' | ''' | ||
== Materials and Methods == | == Materials and Methods == | ||
''' | ''' | ||
| Line 44: | Line 47: | ||
== Experimental Validation == | == Experimental Validation == | ||
''' | ''' | ||
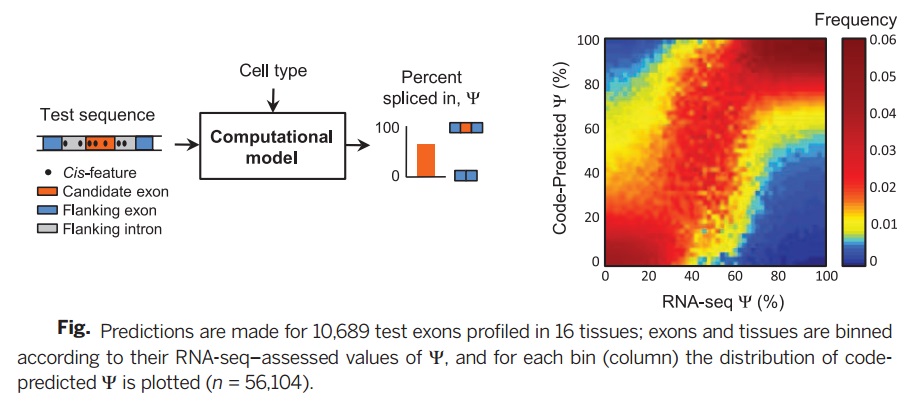
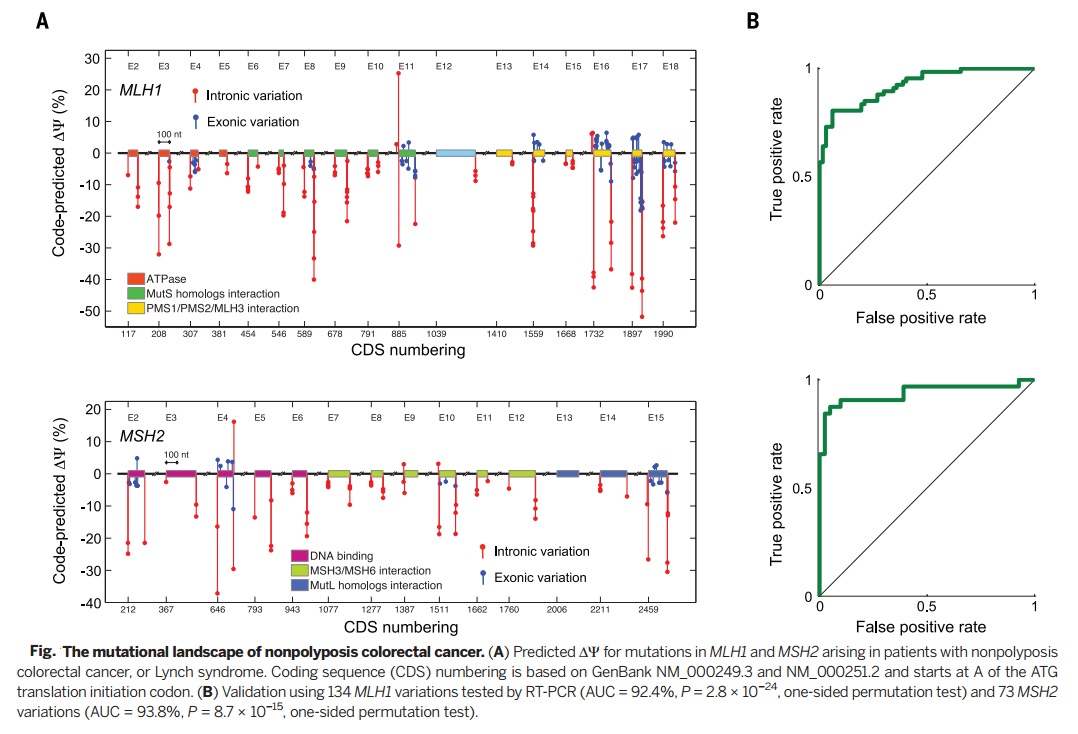
To check the accuracy of the suggested splicing regulatory model, in this research, experimental results of several data bases are used including RNA-seq data, ET-PCR data, RNA binding protein affinity data, splicing factor knockdown data, and phenotypic/genotypic data. | To check the accuracy of the suggested splicing regulatory model, in this research, experimental results of several data bases are used including RNA-seq data, ET-PCR data, RNA binding protein affinity data, splicing factor knockdown data, and phenotypic/genotypic data. | ||
[[File:Stat2.jpg]] | |||
<br /> | |||
[[File:Stat6.jpg]] | |||
''' | ''' | ||
== Genome- | |||
== Genome-wide Analysis == | |||
''' | ''' | ||
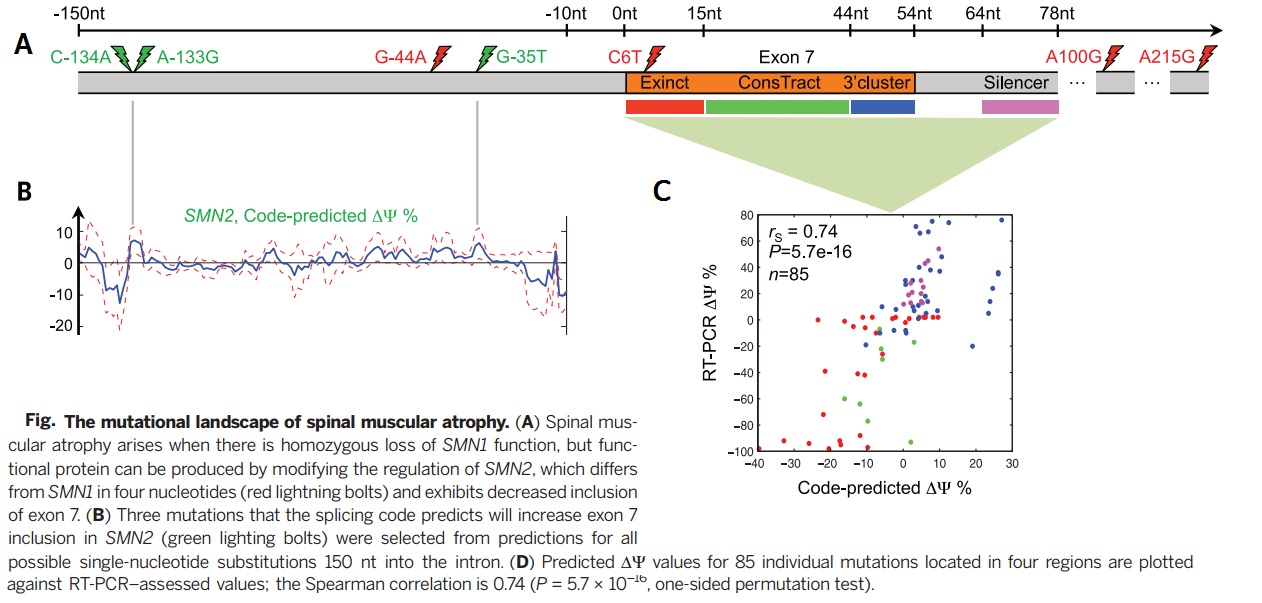
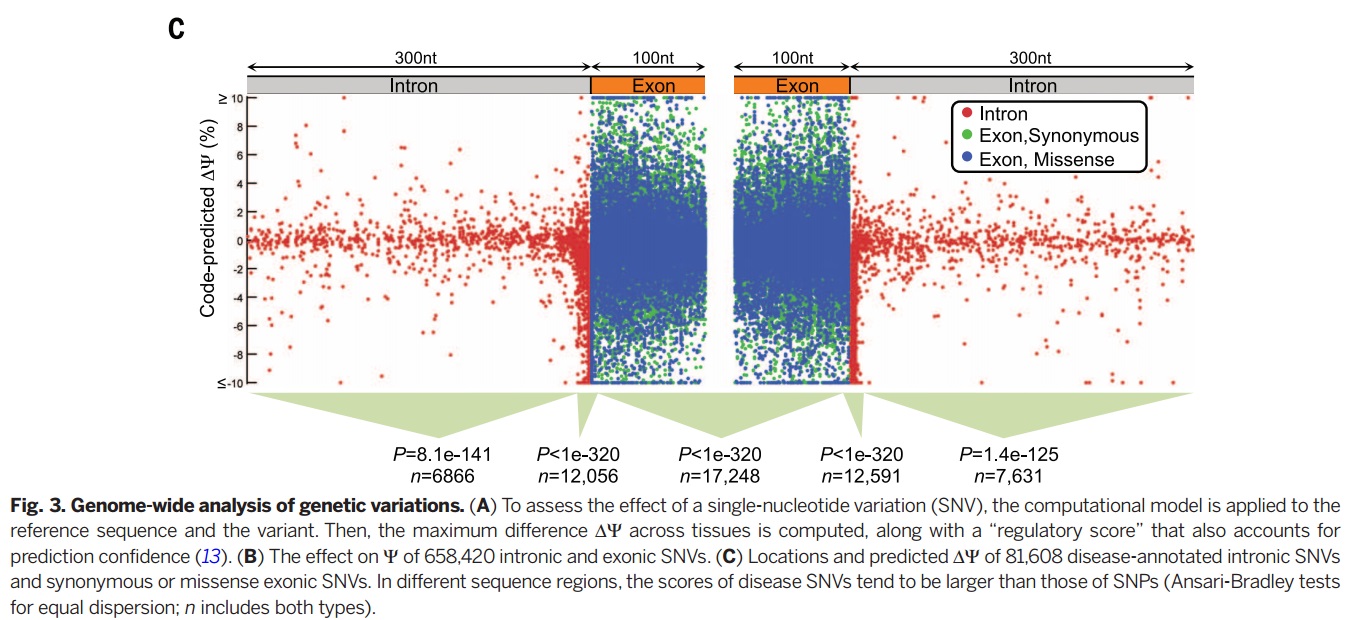
As an important implications of genetic variation of splicing regulation, 658420 SNVs mapped to exonic and intronic sequences. Then the effect of each SNV on splicing regulation scored by applying the regulatory model of finding the largest value of the difference in predicted splicing level <math>\nabla \psi</math> across tissues. | As an important implications of genetic variation of splicing regulation, 658420 SNVs mapped to exonic and intronic sequences. Then the effect of each SNV on splicing regulation scored by applying the regulatory model of finding the largest value of the difference in predicted splicing level <math>\nabla \psi</math> across tissues. | ||
[[File:Stat5.jpg]] | |||
[[File:Stat8.jpg]] | |||
''' | |||
== Conclusion == | == Conclusion == | ||
''' | ''' | ||
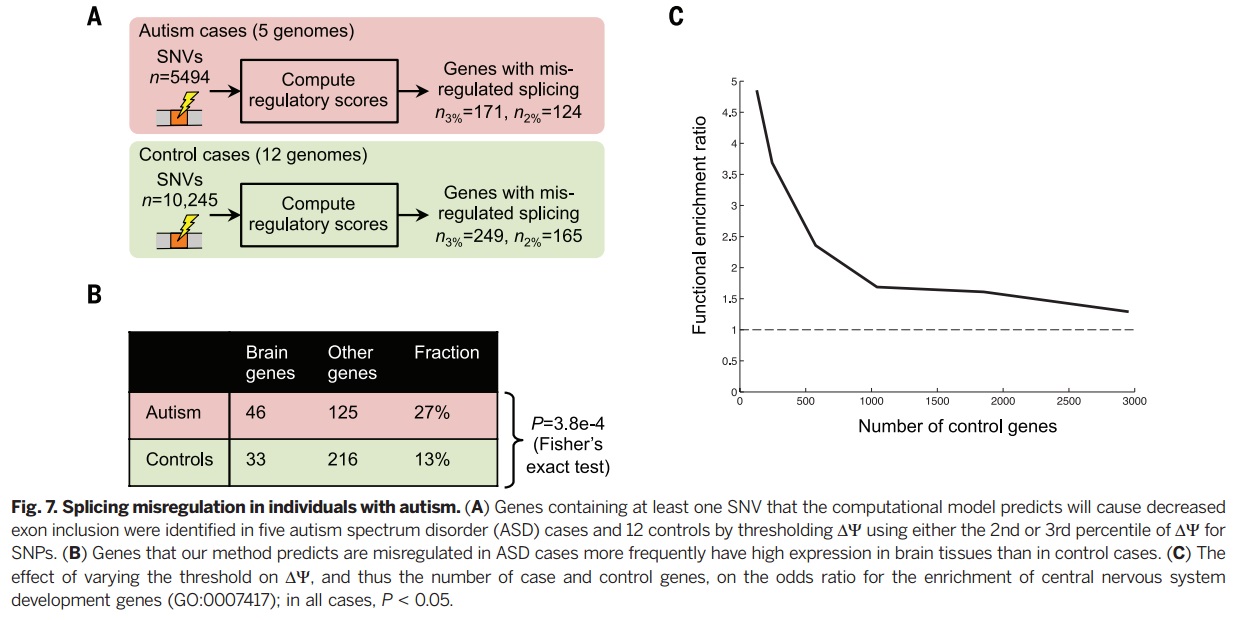
The method introduced in this paper represents a technique for disease-causing variants classification and for aberrant splicing malignancies. This computational model was trained to predict DNA sequence splicing in the absence of disease annotations or other existing population data and thus can be compared as a naive approach to the experimental data. Thus this model provides a method to understand the genetic basis of various diseases. | The method introduced in this paper represents a technique for disease-causing variants classification and for aberrant splicing malignancies. This computational model was trained to predict DNA sequence splicing in the absence of disease annotations or other existing population data and thus can be compared as a naive approach to the experimental data. Thus this model provides a method to understand the genetic basis of various diseases. This technique is able to accurately classify disease-causing variants and provides insights into the role of aberrant splicing in disease. This model predicts substantial and unexpected aberrant splicing due to variants within introns and exons, including those far from the splice site. | ||
We know there are several practical considerations when using Bayes Neural networks. For instance difficulty to speed up and scale up to a large number of hidden variables because of relying on methods like MCMC it is. Leung et al <ref> | |||
Leung M, Deep learning of the tissue-regulated splicing code, Bioiformatics 30, 2014. | |||
</ref>. proposed an architecture that can have thousands of hidden units with multiple non-linear layers and millions of model parameters. | |||
[[File:Stat7.jpg]] | |||
''' | ''' | ||
== References == | == References == | ||
''' | ''' | ||
Latest revision as of 09:46, 30 August 2017
Genetic Application of Deep Learning
This paper presentation is based on the paper [Hui Y. Xiong1 et al, Science 347, 2015] which reveals the importance of deep learning methods in genetic study of disease while using different types of machine-learning approaches would enable us to precise annotation mechanism. These techniques have been done for a wide variety of disease including different cancers which has led to important achievements in mutation-driven splicing. t reach to this goal, various intronic and exonic disease mutations have taken into account to detect variants of mutations. This procedure should enable us to prognosis, diagnosis, and/or control a wide variety of diseases.
Introduction
It has been a while since whole-genome sequencing been used to detect the source of disease or unwanted malignancies genetically. The idea is to find a hierarchy of mutations tending to such diseases by looking at alterations via genetic variations in the genome and particularly when they occur outside of those domains in which protein-coding happens. In the present paper, a computational method is given to detect those genetic variants which influence RNA splicing. RNA splicing is a modification of pre-messenger RNA (pre-mRNA) when introns are removed and makes the exons joined. Any type of interruptions on this important step of gene expression would lead to various kind of disease such as cancers and neurological disorders.
Rationale
Deep learning algorithm is used to construct a computational model in which DNA sequences are inputs to predict splicing in human textures. In this model, test variants up to 300 nucleotides into an intron, can then be used to derive a score for variant alterations for splicing. The model is not biased by existing disease annotations or population data and was derived in such a way that it can be used to study diverse diseases and disorders and to determine the consequences of common, rare, and even spontaneous variants.
Materials and Methods
The human splicing regulatory model is analyzed by Baysian machine learning method. 10,698 cassette exons has considered in this study as a training case. The goal is to maximize an information-theoretic code quality measure [math]\displaystyle{ CQ=\sum_e \sum_t D_{KL} (q_{t,e} | r_t ) - D_{KL} (q_{t,e} | p_{t,e} ) }[/math] where [math]\displaystyle{ q_{t,e} }[/math] is the target splicing pattern for exon in tissue t, [math]\displaystyle{ r_t }[/math] is the optimized guesser's prediction ignoring possible RNA features, [math]\displaystyle{ p_{t,e} }[/math] is the non-trained regulatory prediction on exons, and [math]\displaystyle{ D_{KL} }[/math] is the Kullback-Leibler between two distributions. CQ is, in fact, a likelihood function of [math]\displaystyle{ p_{t,e} }[/math].
The structure of each model is a two-layer neural network of units which are sigmoidal hidden within a considered tissue. In our special case study, nonlinear and texture-dependent correlation between the RNA features and the splicing has considered. In such a model, RNA features provide the inputs to 30 hidden variables at most. Each hidden variable is a sigmoidal non-linearity of its corresponding input. Then by applying a softmax function, the non-linear hidden variable are used to prepare the prediction. Moreover, tissues are also trained jointly as disjoint output units.
Regarding the complexity of this approach, considering maximum likelihood learning method an overfitting is done for each model. The main learning algorithm applied in this paper are from <ref> Xiong H.Y. et al, Baysian Prediction of tissue-regulated splicing using RNA sequence and cellular context, Bioiformation 27, pp. 2554-2562, 2011. </ref>. As a generalization of logistic regression, the multinomial regression model has considered linear in log odds ratio domain and without hidden variables. Then the model is trained by taking into account the same objective function, RNA features, splicing patterns, and partitioning the dataset as the Baysian neutral network described in above.
Experimental Validation
To check the accuracy of the suggested splicing regulatory model, in this research, experimental results of several data bases are used including RNA-seq data, ET-PCR data, RNA binding protein affinity data, splicing factor knockdown data, and phenotypic/genotypic data.
Genome-wide Analysis
As an important implications of genetic variation of splicing regulation, 658420 SNVs mapped to exonic and intronic sequences. Then the effect of each SNV on splicing regulation scored by applying the regulatory model of finding the largest value of the difference in predicted splicing level [math]\displaystyle{ \nabla \psi }[/math] across tissues.
Conclusion
The method introduced in this paper represents a technique for disease-causing variants classification and for aberrant splicing malignancies. This computational model was trained to predict DNA sequence splicing in the absence of disease annotations or other existing population data and thus can be compared as a naive approach to the experimental data. Thus this model provides a method to understand the genetic basis of various diseases. This technique is able to accurately classify disease-causing variants and provides insights into the role of aberrant splicing in disease. This model predicts substantial and unexpected aberrant splicing due to variants within introns and exons, including those far from the splice site. We know there are several practical considerations when using Bayes Neural networks. For instance difficulty to speed up and scale up to a large number of hidden variables because of relying on methods like MCMC it is. Leung et al <ref> Leung M, Deep learning of the tissue-regulated splicing code, Bioiformatics 30, 2014. </ref>. proposed an architecture that can have thousands of hidden units with multiple non-linear layers and millions of model parameters.
References
[1] Hui Y. Xiong1 et al, The human splicing code reveals new insights into the genetic determinants of disease, Science 347, 2015.
[2] Xiong H.Y. et al, Baysian Prediction of tissue-regulated splicing using RNA sequence and cellular context, Bioiformation 27, pp. 2554-2562, 2011.