stat341f11: Difference between revisions
m (Conversion script moved page Stat341f11 to stat341f11: Converting page titles to lowercase) |
|||
| (762 intermediate revisions by 33 users not shown) | |||
| Line 1: | Line 1: | ||
Please contribute to the discussion of splitting up this page into multiple pages on the [[{{TALKPAGENAME}}|talk page]]. | |||
==[[signupformStat341F11| Editor Sign Up]]== | ==[[signupformStat341F11| Editor Sign Up]]== | ||
==Sampling - | ==Notation== | ||
The following guidelines on notation were posted on the Wiki Course Note page for [[Stat946f11|STAT 946]]. Add to them as necessary for consistent notation on this page. | |||
Capital letters will be used to denote random variables and lower case letters denote observations for those random variables: | |||
* <math>\{X_1,\ X_2,\ \dots,\ X_n\}</math> random variables | |||
* <math>\{x_1,\ x_2,\ \dots,\ x_n\}</math> observations of the random variables | |||
The joint ''probability mass function'' can be written as: | |||
<center><math> P( X_1 = x_1, X_2 = x_2, \dots, X_n = x_n )</math></center> | |||
or as shorthand, we can write this as <math>p( x_1, x_2, \dots, x_n )</math>. In these notes both types of notation will be used. | |||
We can also define a set of random variables <math>X_Q</math> where <math>Q</math> represents a set of subscripts. | |||
==Sampling - September 20, 2011== | |||
The meaning of sampling is to generate data points or numbers such that these data follow a certain distribution.<br /> | The meaning of sampling is to generate data points or numbers such that these data follow a certain distribution.<br /> | ||
i.e. From <math>x \sim~f(x)</math> sample <math>\,x_{1}, x_{2}, ..., x_{1000}</math> | i.e. From <math>x \sim~f(x)</math> sample <math>\,x_{1}, x_{2}, ..., x_{1000}</math> | ||
In practice, it maybe difficult to find the joint distribution of random variables. | In practice, it maybe difficult to find the joint distribution of random variables. We will explore different methods for simulating random variables, and how to draw conclusions using the simulated data. | ||
===Sampling from Uniform Distribution=== | |||
Computers cannot generate random numbers as they are deterministic; however they can produce pseudo random numbers using algorithms. Generated numbers mimic the properties of random numbers but they are never truly random. One famous algorithm that is considered highly reliable is the Mersenne twister[http://en.wikipedia.org/wiki/Mersenne_twister], which generates random numbers in an almost uniform distribution. | |||
====Multiplicative Congruential==== | ====Multiplicative Congruential==== | ||
*involves | *involves four parameters: integers <math>\,a, b, m</math>, and an initial value <math>\,x_0</math> which we call the seed | ||
*a sequence of integers is defined as | *a sequence of integers is defined as | ||
:<math>x_{k+1} \equiv (ax_{k} + b) \mod{m}</math> | :<math>x_{k+1} \equiv (ax_{k} + b) \mod{m}</math> | ||
| Line 21: | Line 39: | ||
<pre> | <pre> | ||
a=13; | a = 13; | ||
b=0; | b = 0; | ||
m=31; | m = 31; | ||
x(1)=1; | x(1) = 1; | ||
for ii = | for ii = 2:1000 | ||
x(ii | x(ii) = mod(a*x(ii-1)+b, m); | ||
end | end | ||
</pre> | </pre> | ||
| Line 42: | Line 60: | ||
hist(x) | hist(x) | ||
</pre> | </pre> | ||
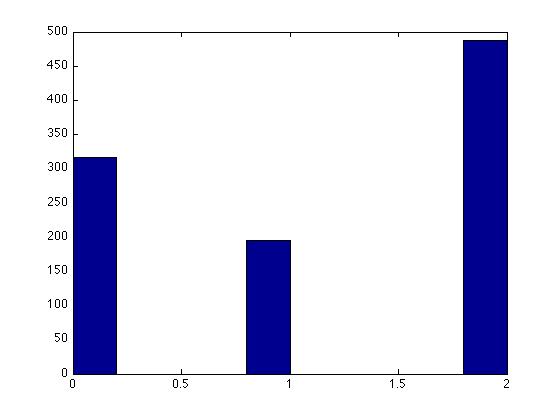
Histogram Output: | |||
[[File:uniform.jpg]] | |||
Facts about this algorithm: | Facts about this algorithm: | ||
*In this example, the first 30 terms in the sequence are a permutation of integers from 1 to 30 and then the sequence repeats itself. | *In this example, the first 30 terms in the sequence are a permutation of integers from 1 to 30 and then the sequence repeats itself. In the general case, this algorithm has a period of m-1. | ||
*Values are between 0 and < | *Values are between <b>0</b> and <b>m-1</b>, inclusive. | ||
*Dividing the numbers by < | *Dividing the numbers by <b> m-1 </b> yields numbers in the interval <b>[0,1]</b>. | ||
*MATLAB's <code>rand</code> function once used this algorithm with < | *MATLAB's <code>rand</code> function once used this algorithm with <b>a= 7<sup>5</sup></b>, <b>b= 0</b>, <b>m= 2<sup>31</sup>-1</b>,for reasons described in Park and Miller's 1988 paper "Random Number Generators: Good Ones are Hard to Find" (available [http://www.firstpr.com.au/dsp/rand31/p1192-park.pdf online]). | ||
*Visual Basic's <code>RND</code> function also used this algorithm with < | *Visual Basic's <code>RND</code> function also used this algorithm with <b>a= 1140671485</b>, <b>b= 12820163</b>, <b>m= 2<sup>24</sup></b>. ([http://support.microsoft.com/kb/231847 Reference]) | ||
===Inverse Transform Method=== | ===Inverse Transform Method=== | ||
This is a basic method for sampling. Theoretically using this method we can generate sample numbers at random from any probability distribution once we know its cumulative distribution function (cdf). | This is a basic method for sampling. Theoretically using this method we can generate sample numbers at random from any probability distribution once we know its cumulative distribution function (cdf). This method is very efficient computationally if the cdf of can be analytically inverted. | ||
====Theorem==== | ====Theorem==== | ||
Take <math>U \sim~ \mathrm{Unif}[0, 1]</math> and let <math>X=F^{-1}(U)</math>. Then X has distribution function <math>F(\cdot)</math>, | Take <math>U \sim~ \mathrm{Unif}[0, 1]</math> and let <math>\ X = F^{-1}(U) </math>. Then <math>\ X</math> has a cumulative distribution function of <math>F(\cdot)</math>, ie. <math>F(x)=P(X \leq x)</math>, where <math>F^{-1}(\cdot)</math> is the inverse of <math>F(\cdot)</math>. | ||
'''Proof''' | '''Proof''' | ||
| Line 81: | Line 101: | ||
*Step 1. Draw <math>U \sim~ \mathrm{Unif}[0, 1]</math> | *Step 1. Draw <math>U \sim~ \mathrm{Unif}[0, 1]</math> | ||
*Step 2. < | *Step 2. <b><i>X=F <sup>−1</sup>(U)</i></b> | ||
'''Example''' | '''Example''' | ||
| Line 87: | Line 107: | ||
Take the exponential distribution for example | Take the exponential distribution for example | ||
:<math>\,f(x)={\lambda}e^{-{\lambda}x}</math> | :<math>\,f(x)={\lambda}e^{-{\lambda}x}</math> | ||
:<math>\,F(x)=\int_0^x {\lambda}e^{-{\lambda}u} du=1-e^{-{\lambda}x}</math> | :<math>\,F(x)=\int_0^x {\lambda}e^{-{\lambda}u} du=[-e^{-{\lambda}u}]_0^x=1-e^{-{\lambda}x}</math> | ||
Let: <math>\,F(x)=y</math> | Let: <math>\,F(x)=y</math> | ||
| Line 98: | Line 118: | ||
*Step 1. Draw <math>U \sim~ \mathrm{Unif}[0, 1]</math> | *Step 1. Draw <math>U \sim~ \mathrm{Unif}[0, 1]</math> | ||
*Step 2. <math>x=\frac{-ln(1-U)}{\lambda}</math> | *Step 2. <math>x=\frac{-ln(1-U)}{\lambda}</math> | ||
Note: If U~Unif[0, 1], then (1 - U) and U have the same distribution. This allows us to slightly simplify step 2 into an alternate form: | |||
*Alternate Step 2. <math>x=\frac{-ln(U)}{\lambda}</math> | |||
'''MATLAB code''' | '''MATLAB code''' | ||
| Line 105: | Line 128: | ||
for ii = 1:1000 | for ii = 1:1000 | ||
u = rand; | u = rand; | ||
x(ii)=-log(1-u)/0.5 | x(ii) = -log(1-u)/0.5; | ||
end | end | ||
hist(x) | hist(x) | ||
</pre> | </pre> | ||
MATLAB result | '''MATLAB result''' | ||
[[File:MATLAB_Exp.jpg|center| | [[File:MATLAB_Exp.jpg|center|700px]] | ||
====Discrete Case==== | ====Discrete Case - September 22, 2011==== | ||
This same technique can be applied to the discrete case. Generate a discrete random variable <math>\,x</math> that has probability mass function <math>\,P(X=x_i)=P_i </math> where <math>\,x_0<x_1<x_2...</math> and <math>\,\sum_i P_i=1</math> | This same technique can be applied to the discrete case. Generate a discrete random variable <math>\,x</math> that has probability mass function <math>\,P(X=x_i)=P_i </math> where <math>\,x_0<x_1<x_2...</math> and <math>\,\sum_i P_i=1</math> | ||
*Step 1. Draw <math>u \sim~ \mathrm{Unif}[0, 1]</math> | *Step 1. Draw <math>u \sim~ \mathrm{Unif}[0, 1]</math> | ||
*Step 2. <math>\,x=x_i</math> if <math>\,F(x_{i-1})<u \leq F(x_i)</math> | *Step 2. <math>\,x=x_i</math> if <math>\,F(x_{i-1})<u \leq F(x_i)</math> | ||
'''Example''' | |||
Let x be a discrete random variable with the following probability mass function: | Let x be a discrete random variable with the following probability mass function: | ||
| Line 139: | Line 159: | ||
F(x) = \begin{cases} | F(x) = \begin{cases} | ||
0 & x < 0 \\ | 0 & x < 0 \\ | ||
0.3 & | 0.3 & x \leq 0 \\ | ||
0.5 & | 0.5 & x \leq 1 \\ | ||
1 & | 1 & x \leq 2 | ||
\end{cases}</math> | \end{cases}</math> | ||
| Line 149: | Line 169: | ||
<pre> | <pre> | ||
Draw U ~ Unif[0,1] | Draw U ~ Unif[0,1] | ||
if U <= 0.3 | |||
return 0 | |||
else if 0.3 < U <= 0.5 | |||
return 1 | |||
else if 0.5 < U <= 1 | |||
return 2 | |||
</pre> | </pre> | ||
| Line 163: | Line 181: | ||
<pre> | <pre> | ||
for ii = 1:1000 | for ii = 1:1000 | ||
u = rand; | |||
if u <= 0.3 | |||
x(ii) = 0; | |||
else if u <= 0.5 | |||
x(ii) = 1; | |||
else | |||
x(ii) = 2; | |||
end | |||
end | end | ||
</pre> | </pre> | ||
Matlab Output: | |||
[[File:Discreteinv.jpg]] | |||
'''Pseudo code for the Discrete Case:''' | '''Pseudo code for the Discrete Case:''' | ||
| Line 180: | Line 201: | ||
1. Draw U ~ Unif [0,1] | 1. Draw U ~ Unif [0,1] | ||
2. If <math> U \leq P_0 </math>, deliver < | 2. If <math> U \leq P_0 </math>, deliver <b><i>X= x<sub>0</sub></i></b> | ||
3. Else if <math> U \leq P_0 + P_1 </math>, deliver < | 3. Else if <math> U \leq P_0 + P_1 </math>, deliver <b><i>X= x<sub>1</sub></i></b> | ||
4. Else If <math> U \leq P_0 +....+ P_k </math>, deliver < | 4. Else If <math> U \leq P_0 +....+ P_k </math>, deliver <b><i>X= x<sub>k</sub></i></b> | ||
====Limitations==== | |||
This method is useful, but it's not practical in many cases because we can't always obtain <math>F</math> or <math> F^{-1} </math> (some functions are not integrable or invertible), and sometimes <math>f(x)</math> cannot be obtained in closed form. Let's look at some examples: | |||
*Continuous case | *Continuous case | ||
If we want to use this method to draw the ''pdf'' of '''normal distribution''', we may find ourselves | If we want to use this method to draw the ''pdf'' of '''normal distribution''', we may find ourselves geting stuck when trying to find its ''cdf''. | ||
The simplest case of '''normal distribution''' is <math>f(x)=\frac{1}{\sqrt{2\pi}}e^{-\frac{x^2}{2}}</math>, | The simplest case of '''normal distribution''' is <math>f(x)=\frac{1}{\sqrt{2\pi}}e^{-\frac{x^2}{2}}</math>, | ||
whose ''cdf'' is <math>F(x)=\frac{1}{\sqrt{2\pi}}\int_{-\infty}^{x}{e^{-\frac{u^2}{2}}}du</math>. This integral cannot be expressed in terms of elementary functions. So evaluating it and then finding the inverse is a very difficult task. | whose ''cdf'' is <math>F(x)=\frac{1}{\sqrt{2\pi}}\int_{-\infty}^{x}{e^{-\frac{u^2}{2}}}du</math>. This integral cannot be expressed in terms of elementary functions. So evaluating it and then finding the inverse is a very difficult task. | ||
| Line 197: | Line 219: | ||
But when n takes on values that are very large, say 50, it is hard to do so. | But when n takes on values that are very large, say 50, it is hard to do so. | ||
===Acceptance/Rejection | ===Acceptance/Rejection Method=== | ||
The aforementioned difficulties of the inverse transform method motivates a sampling method that does not require analytically calculating cdf's and their inverses, which is the acceptance/rejection sampling method. Here, <math> f(x)</math> is approximated by another function, say <math>g(x)</math>, with the idea being that <math>g(x)</math> is a "nicer" function to work with than <math>f(x)</math>. | The aforementioned difficulties of the inverse transform method motivates a sampling method that does not require analytically calculating cdf's and their inverses, which is the acceptance/rejection sampling method. Here, <math> \displaystyle f(x)</math> is approximated by another function, say <math>\displaystyle g(x)</math>, with the idea being that <math>\displaystyle g(x)</math> is a "nicer" function to work with than <math>\displaystyle f(x)</math>. | ||
Suppose we assume the following: | Suppose we assume the following: | ||
1. There exists another distribution <math>g(x)</math> that is easier to work with and that you know how to sample from, and | 1. There exists another distribution <math>\displaystyle g(x)</math> that is easier to work with and that you know how to sample from, and | ||
2. There exists a constant c such that <math>f(x) \leq c \cdot g(x)</math> for all x | 2. There exists a constant c such that <math>f(x) \leq c \cdot g(x)</math> for all x | ||
Under these assumptions, we can sample from <math>f(x)</math> by sampling from <math>g(x)</math> | Under these assumptions, we can sample from <math>\displaystyle f(x)</math> by sampling from <math>\displaystyle g(x)</math> | ||
====General Idea==== | ====General Idea==== | ||
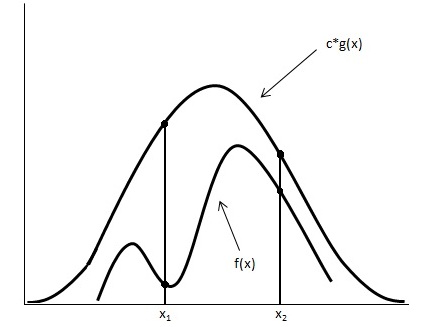
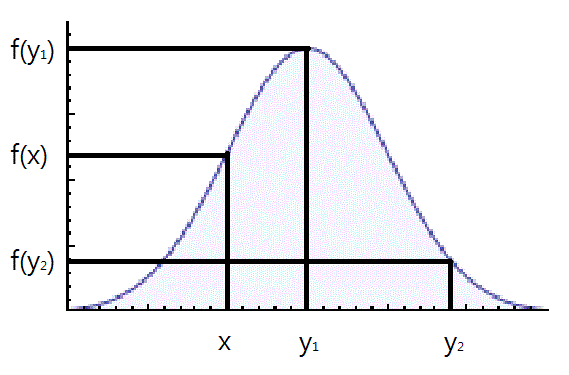
Looking at the image below we have graphed <math> c \cdot g(x) </math> and <math>f(x)</math>. | Looking at the image below we have graphed <math> c \cdot g(x) </math> and <math>\displaystyle f(x)</math>. | ||
[[File:Graph_updated.jpg]] | [[File:Graph_updated.jpg]] | ||
Using the acceptance/rejection method we will accept some of the points from <math>g(x)</math> and reject some of the points from <math> g(x)</math>. The points that will be accepted from <math>g(x)</math> will have a distribution similar to <math>f(x)</math>. We can see from the image that the values around <math>x_1</math> will be sampled more often under <math>c \cdot g(x)</math> than under <math>f(x)</math>, so we will have to reject more samples taken at x<sub>1</sub>. Around <math>x_2</math> the number of samples that are drawn and the number of samples we need are much closer, so we accept more samples that we get at <math>x_2</math> | Using the acceptance/rejection method we will accept some of the points from <math>\displaystyle g(x)</math> and reject some of the points from <math>\displaystyle g(x)</math>. The points that will be accepted from <math>\displaystyle g(x)</math> will have a distribution similar to <math>\displaystyle f(x)</math>. We can see from the image that the values around <math>\displaystyle x_1</math> will be sampled more often under <math>c \cdot g(x)</math> than under <math>\displaystyle f(x)</math>, so we will have to reject more samples taken at x<sub>1</sub>. Around <math>\displaystyle x_2</math> the number of samples that are drawn and the number of samples we need are much closer, so we accept more samples that we get at <math>\displaystyle x_2</math>. We should remember to take these considerations into account when evaluating the efficiency of a given acceptance-rejection method. Rejecting a high proportion of samples ultimately leaves us with a longer time until we retrieve our desired distribution. We should question whether a better function <math> g(x) </math> can be chosen in our situation. | ||
====Procedure==== | ====Procedure==== | ||
| Line 224: | Line 247: | ||
3. If <math> U \leq \frac{f(y)}{c \cdot g(y)}</math> then x=y; else return to 1 | 3. If <math> U \leq \frac{f(y)}{c \cdot g(y)}</math> then x=y; else return to 1 | ||
Note that the choice of <math> c </math> plays an important role in the efficiency of the algorithm. We want <math> c \cdot g(x) </math> to be "tightly fit" over <math> f(x) </math> to increase the probability of accepting points, and therefore reducing the number of sampling attempts. Mathematically, we want to minimize <math> c </math> such that <math>f(x) \leq c \cdot g(x) \ \forall x</math>. We do this by setting | |||
<math> \frac{d}{dx}(\frac{f(x)}{g(x)}) = 0 </math>, solving for a maximum point <math> x_0 </math> and setting <math> c = \frac{f(x_0)}{g(x_0)}. </math> | |||
====Proof==== | |||
Mathematically, we need to show that the sample points given that they are accepted have a distribution of f(x). | |||
<math>\begin{align} P(y|accepted) &= \frac{P(y, accepted)}{P(accepted)} \\ | |||
&= \frac{P(accepted|y) P(y)}{P(accepted)}\end{align} </math> (Bayes' Rule) | |||
<math>\displaystyle P(y) = g(y)</math> | |||
<math>P(accepted|y) =P(u\leq \frac{f(y)}{c \cdot g(y)}) =\frac{f(y)}{c \cdot g(y)} </math>,where u ~ Unif [0,1] | |||
<math>P(accepted) = \sum P(accepted|y)\cdot P(y)=\int^{}_y \frac{f(s)}{c \cdot g(s)}g(s) ds=\int^{}_y \frac{f(s)}{c} ds=\frac{1}{c} \cdot\int^{}_y f(s) ds=\frac{1}{c}</math> | |||
So, | |||
<math> P(y|accepted) = \frac{ \frac {f(y)}{c \cdot g(y)} \cdot g(y)}{\frac{1}{c}} =f(y) </math> | |||
====Continuous Case==== | |||
'''Example''' | '''Example''' | ||
| Line 233: | Line 280: | ||
In general: | In general: | ||
<math>Beta(\alpha, \beta) = \frac{\Gamma (\alpha + \beta)}{\Gamma(\alpha)\Gamma(\beta)}</math> <math>\displaystyle x^{\alpha-1}</math> <math>\displaystyle(1-x)^{\beta-1}</math>, <math>\displaystyle 0<x<1</math> | |||
Note: <math>\Gamma(n) = (n-1)!</math> if n is a positive integer | Note: <math>\!\Gamma(n) = (n-1)!</math> if n is a positive integer | ||
<math>\begin{align} f(x) &= Beta(2,1) \\ | <math>\begin{align} f(x) &= Beta(2,1) \\ | ||
| Line 242: | Line 289: | ||
&= 2x \end{align}</math> | &= 2x \end{align}</math> | ||
We want to choose <math>g(x)</math> that is easy to sample from. So we choose <math>g(x)</math> to be uniform distribution. | We want to choose <math>\displaystyle g(x)</math> that is easy to sample from. So we choose <math>\displaystyle g(x)</math> to be uniform distribution on <math>\ (0,1)</math> since that is the domain for the Beta function. | ||
We now want a constant c such that <math>f(x) \leq c \cdot g(x) </math> for all x from (0,1) | We now want a constant c such that <math>f(x) \leq c \cdot g(x) </math> for all x from Unif(0,1) | ||
So,<br /> | So,<br /> | ||
<math>c \geq \frac{f(x)}{g(x)}</math>, for all x from (0,1) | <math>c \geq \frac{f(x)}{g(x)}</math>, for all x from (0,1) | ||
| Line 293: | Line 340: | ||
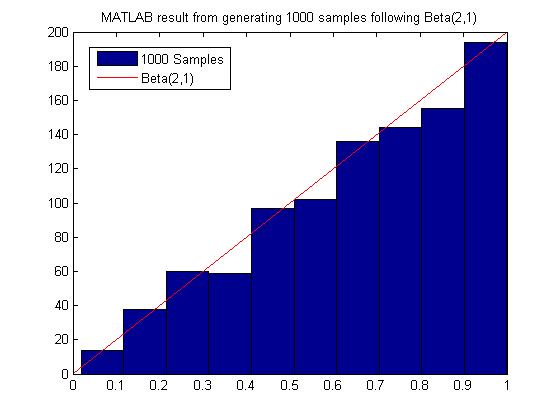
[[File:MATLAB_Beta.jpg]] | [[File:MATLAB_Beta.jpg]] | ||
= | ====Discrete Example==== | ||
===Discrete Example=== | |||
Generate random variables according to the p.m.f: | Generate random variables according to the p.m.f: | ||
| Line 359: | Line 382: | ||
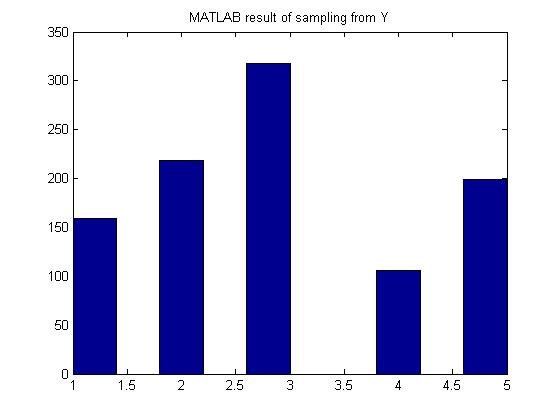
[[File:MATLAB_Y.jpg]] | [[File:MATLAB_Y.jpg]] | ||
== | ====Limitations==== | ||
===Sampling From Gamma=== | Most of the time we have to sample many more points from g(x) before we can obtain an acceptable amount of samples from f(x), hence this method may not be computationally efficient. It depends on our choice of g(x). For example, in the example above to sample from Beta(2,1), we need roughly 2000 samples from g(X) to get 1000 acceptable samples of f(x). | ||
In addition, in situations where a g(x) function is chosen and used, there can be a discrepancy between the functional behaviors of f(x) and g(x) that render this method unreliable. For example, given the normal distribution function as g(x) and a function of f(x) with a "fat" mid-section and "thin tails", this method becomes useless as more points near the two ends of f(x) will be rejected, resulting in a tedious and overwhelming number of sampling points having to be sampled due to the high rejection rate of such a method. | |||
===Sampling From Gamma and Normal Distribution - September 27, 2011=== | |||
====Sampling From Gamma==== | |||
'''Gamma Distribution''' | |||
The Gamma function is written as <math>X \sim~ Gamma (t, \lambda) </math> | The Gamma function is written as <math>X \sim~ Gamma (t, \lambda) </math> | ||
<math> F(x) = \int_{0}^{\lambda x} \frac{e^{-y}y^{t-1}}{(t-1)!} dy </math> | :<math> F(x) = \int_{0}^{\lambda x} \frac{e^{-y}y^{t-1}}{(t-1)!} dy </math> | ||
If you have t samples of the exponential distribution,<br> | If you have t samples of the exponential distribution,<br> | ||
<br> <math> \begin{align} | <br> <math> \begin{align} X_1 \sim~ Exp(\lambda)\\ \vdots \\ X_t \sim~ Exp(\lambda) \end{align} | ||
</math> | </math> | ||
The sum of these t samples has a gamma distribution, | The sum of these t samples has a gamma distribution, | ||
<math> | :<math> X_1+X_2+ ... + X_t \sim~ Gamma (t, \lambda) </math><br> | ||
<math> \sum_{i=1}^{t} | :<math> \sum_{i=1}^{t} X_i \sim~ Gamma (t, \lambda) </math> where <math>X_i \sim~Exp(\lambda)</math><br> | ||
We can sample the exponential distribution using the inverse transform method from previous class,<br> | '''Method''' <br> | ||
<math>\,f(x)={\lambda}e^{-{\lambda}x}</math><br> | Suppose we want to sample <math>\ k </math> points from <math>\ Gamma (t, \lambda) </math>. <br> | ||
<math>\,F^{-1}(u)=\frac{-ln(1-u)}{\lambda}</math><br> | We can sample the exponential distribution using the inverse transform method from the previous class,<br> | ||
<math>\,F^{-1}(u)=\frac{-ln(u)}{\lambda}</math> 1 - u is the same as x since <math>U \sim~ unif [0,1] </math><br> | :<math>\,f(x)={\lambda}e^{-{\lambda}x}</math><br> | ||
<math> \begin{align} \frac{-ln(u_1)}{\lambda} - \frac{ln(u_2)}{\lambda} - ... - \frac{ln(u_t)}{\lambda} = x_1\\ \vdots \\ \frac{-ln(u_1)}{\lambda} - \frac{ln(u_2)}{\lambda} - ... - \frac{ln(u_t)}{\lambda} = | :<math>\,F^{-1}(u)=\frac{-ln(1-u)}{\lambda}</math><br> | ||
</math><br> | :<math>\,F^{-1}(u)=\frac{-ln(u)}{\lambda}</math> | ||
<math> \frac {-\sum_{i=1}^{t} ln(u_i)}{\lambda} = x</math> | (1 - u) is the same as x since <math>U \sim~ unif [0,1] </math><br> | ||
:<math> \begin{align} \frac{-ln(u_1)}{\lambda} - \frac{ln(u_2)}{\lambda} - ... - \frac{ln(u_t)}{\lambda} = x_1\\ \vdots \\ \frac{-ln(u_1)}{\lambda} - \frac{ln(u_2)}{\lambda} - ... - \frac{ln(u_t)}{\lambda} = x_k \end{align} | |||
:</math><br> | |||
:<math> \frac {-\sum_{i=1}^{t} ln(u_i)}{\lambda} = x</math> | |||
MATLAB code for a Gamma(1 | '''MATLAB code''' for a Gamma(3,1) is | ||
<pre> | <pre> | ||
| Line 393: | Line 427: | ||
And the Histogram of X follows a Gamma distribution with long tail: | And the Histogram of X follows a Gamma distribution with long tail: | ||
[[File:Hist.PNG]] | [[File:Hist.PNG|center|500px]] | ||
We can improve the quality of histogram by adjusting the Matlab code to include the number of bins we want: hist(x, number_of_bins) | |||
<pre> | |||
x = sum(-log(rand(20000,3)),2); | |||
hist(x,40) | |||
</pre> | |||
[[File:untitled.jpg|center|500px]] | |||
''' R code''' for a Gamma(3,1) is | |||
<pre> | <pre> | ||
a<-apply(-log(matrix(runif(3000),nrow=1000)),1,sum); | a<-apply(-log(matrix(runif(3000),nrow=1000)),1,sum); | ||
hist(a); | hist(a); | ||
</pre> | </pre> | ||
Histogram: | |||
[[File:hist_gamma.png]] | [[File:hist_gamma.png|center|500px]] | ||
Here is another histogram of Gamma coding with R | Here is another histogram of Gamma coding with R | ||
| Line 412: | Line 454: | ||
rug(jitter(a)); | rug(jitter(a)); | ||
</pre> | </pre> | ||
[[File:hist_gamma_2.png]] | [[File:hist_gamma_2.png|center|500px]] | ||
===Box-Muller Transform=== | ====Sampling from Normal Distribution using Box-Muller Transform - September 29, 2011==== | ||
=====Procedure===== | |||
==== | # Generate <math>\displaystyle u_1</math> and <math>\displaystyle u_2</math>, two values sampled from a uniform distribution between 0 and 1. | ||
# Set <math>\displaystyle R^2 = -2log(u_1)</math> so that <math>\displaystyle R^2</math> is exponential with mean 2 <br> Set <math>\!\theta = 2*\pi*u_2</math> so that <math>\!\theta</math> ~ <math>\ Unif[0, 2\pi] </math> | |||
# Set <math>\displaystyle X = R cos(\theta)</math> <br> Set <math>\displaystyle Y = R sin(\theta)</math> | |||
=====Justification===== | |||
Suppose we have X ~ N(0, 1) and Y ~ N(0, 1) where X and Y are independent normal random variables. The relative probability density function of these two random variables using Cartesian coordinates is: | |||
<math> f(X, Y) dxdy= f(X) f(Y) dxdy= \frac{1}{\sqrt{2\pi}}e^{-x^2/2} \frac{1}{\sqrt{2\pi}}e^{-y^2/2} dxdy= \frac{1}{2\pi}e^{-(x^2+y^2)/2}dxdy </math> <br> | |||
In polar coordinates <math>\displaystyle R^2 = x^2 + y^2</math>, so the relative probability density function of these two random variables using polar coordinates is: | |||
In polar coordinates <math>R^2 = x^2 + y^2</math>, so the relative probability density function of these two random variables using polar coordinates is: | |||
<math> f(R, \theta) = \frac{1}{2\pi}e^{-R^2/2} </math> <br> | <math> f(R, \theta) = \frac{1}{2\pi}e^{-R^2/2} </math> <br> | ||
If we have <math>R^2 | If we have <math>\displaystyle R^2 \sim exp(1/2)</math> and <math>\!\theta \sim unif[0, 2\pi]</math> we get an equivalent relative probability density function. Notice that when doing a two by two linear transformation, the determinant of the Jacobian needs to be included in the change of variable formula where: | ||
<math> |J|=\left|\frac{\partial(x,y)}{\partial(R,\theta)}\right|= \left|\begin{matrix}\frac{\partial x}{\partial R}&\frac{\partial x}{\partial \theta}\\\frac{\partial y}{\partial R}&\frac{\partial y}{\partial \theta}\end{matrix}\right|=R </math> <br> | |||
<math> f( | <math> f(X, Y) dxdy = f(R, \theta)|J|dRd\theta = \frac{1}{2\pi}e^{-R^2/2}R dRd\theta= \frac{1}{4\pi}e^{-\frac{s}{2}} dSd\theta </math> <br>where <math> S=R^2. </math> <br> | ||
Therefore we can generate a point in polar coordinates using the uniform and exponential distributions, then convert the point to Cartesian coordinates and the resulting X and Y values will be equivalent to samples generated from N(0, 1). | Therefore we can generate a point in polar coordinates using the uniform and exponential distributions, then convert the point to Cartesian coordinates and the resulting X and Y values will be equivalent to samples generated from N(0, 1). | ||
'''MATLAB code''' | |||
In MatLab this algorithm can be implemented with the following code, which generates 20,000 samples from N(0, 1): | In MatLab this algorithm can be implemented with the following code, which generates 20,000 samples from N(0, 1): | ||
| Line 486: | Line 501: | ||
In one execution of this script, the following histogram for x was generated: | In one execution of this script, the following histogram for x was generated: | ||
[[File:Hist standard normal.jpg]] | [[File:Hist standard normal.jpg|center|500px]] | ||
===Non-Standard Normal Distributions=== | =====Non-Standard Normal Distributions===== | ||
'''Example 1: Single-variate Normal''' | |||
If X ~ Norm(0, 1) then (a + bX) has a normal distribution with a mean of <math>\displaystyle a</math> and a standard deviation of <math>\displaystyle b</math> (which is equivalent to a variance of <math>\displaystyle b^2</math>). Using this information with the Box-Muller transform, we can generate values sampled from some random variable <math>\displaystyle Y\sim N(a,b^2) </math> for arbitrary values of <math>\displaystyle a,b</math>. | |||
# Generate a sample u from Norm(0, 1) using the Box-Muller transform. | |||
# Set v = a + bu. | |||
The values for v generated in this way will be equivalent to | The values for v generated in this way will be equivalent to sample from a <math>\displaystyle N(a, b^2)</math>distribution. We can modify the MatLab code used in the last section to demonstrate this. We just need to add one line before we generate the histogram: | ||
<pre> | |||
v = a + b * x; | |||
</pre> | |||
For instance, this is the histogram generated when b = 15, a = 125: | For instance, this is the histogram generated when b = 15, a = 125: | ||
[[File:Hist normal.jpg]] | [[File:Hist normal.jpg|center|500]] | ||
'''Example 2: Multi-variate Normal''' | |||
// In | The Box-Muller method can be extended to higher dimensions to generate multivariate normals. The objects generated will be nx1 vectors, and their variance will be described by nxn covariance matrices. | ||
<math>\mathbf{z} = N(\mathbf{u}, \Sigma)</math> defines the n by 1 vector <math>\mathbf{z}</math> such that: | |||
* <math>\displaystyle u_i</math> is the average of <math>\displaystyle z_i</math> | |||
* <math>\!\Sigma_{ii}</math> is the variance of <math>\displaystyle z_i</math> | |||
* <math>\!\Sigma_{ij}</math> is the co-variance of <math>\displaystyle z_i</math> and <math>\displaystyle z_j</math> | |||
If <math>\displaystyle z_1, z_2, ..., z_d</math> are normal variables with mean 0 and variance 1, then the vector <math>\displaystyle (z_1, z_2,..., z_d) </math> has mean 0 and variance <math>\!I</math>, where 0 is the zero vector and <math>\!I</math> is the identity matrix. This fact suggests that the method for generating a multivariate normal is to generate each component individually as single normal variables. | |||
The mean and the covariance matrix of a multivariate normal distribution can be adjusted in ways analogous to the single variable case. If <math>\mathbf{z} \sim N(0,I)</math>, then <math>\Sigma^{1/2}\mathbf{z}+\mu \sim N(\mu,\Sigma)</math>. Note here that the covariance matrix is symmetric and nonnegative, so its square root should always exist. | |||
We can compute <math>\mathbf{z}</math> in the following way: | |||
# Generate an n by 1 vector <math>\mathbf{x} = \begin{bmatrix}x_{1} & x_{2} & ... & x_{n}\end{bmatrix}</math> where <math>x_{i}</math> ~ Norm(0, 1) using the Box-Muller transform. | |||
# Calculate <math>\!\Sigma^{1/2}</math> using singular value decomposition. | |||
# Set <math>\mathbf{z} = \Sigma^{1/2} \mathbf{x} + \mathbf{u}</math>. | |||
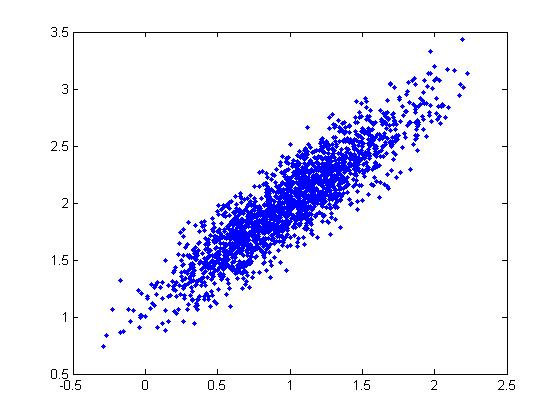
The following MatLab code provides an example, where a scatter plot of 10000 random points is generated. In this case x and y have a co-variance of 0.9 - a very strong positive correlation. | |||
<pre> | <pre> | ||
| Line 524: | Line 560: | ||
root_E = u * (s ^ (1 / 2)); | root_E = u * (s ^ (1 / 2)); | ||
z = (root_E * [x y]') | z = (root_E * [x y]'); | ||
z(: | z(1,:) = z(1,:) + 0; | ||
z(: | z(2,:) = z(2,:) + -3; | ||
scatter(z(: | scatter(z(1,:), z(2,:)) | ||
</pre> | </pre> | ||
This code generated the following scatter plot: | This code generated the following scatter plot: | ||
[[File:scatter covar.jpg]] | [[File:scatter covar.jpg|center|500px]] | ||
In Matlab, we can also use the function "sqrtm()" or "chol()" (Cholesky Decomposition) to calculate square root of a matrix directly. Note that the resulting root matrices may be different but this does materially affect the simulation. | |||
Here is an example: | |||
<pre> | |||
E = [1, 0.9; 0.9, 1]; | |||
r1 = sqrtm(E); | |||
r2 = chol(E); | |||
</pre> | |||
R code for a multivariate normal distribution: | |||
<pre> | |||
n=10000; | |||
r2<--2*log(runif(n)); | |||
theta<-2*pi*(runif(n)); | |||
x<-sqrt(r2)*cos(theta); | |||
y<-sqrt(r2)*sin(theta); | |||
a<-matrix(c(x,y),nrow=n,byrow=F); | |||
e<-matrix(c(1,.9,09,1),nrow=2,byrow=T); | |||
svde<-svd(e); | |||
root_e<-svde$u %*% diag(svde$d)^1/2; | |||
z<-t(root_e %*%t(a)); | |||
z[,1]=z[,1]+5; | |||
z[,2]=z[,2]+ -8; | |||
par(pch=19); | |||
plot(z,col=rgb(1,0,0,alpha=0.06)) | |||
</pre> | |||
[[File:m_normal.png|center|500px]] | |||
=====Remarks===== | |||
MATLAB's randn function uses the ziggurat method to generate normal distributed samples. It is an efficient rejection method based on covering the probability density function with a set of horizontal rectangles so as to obtain points within each rectangle. It is reported that a 800 MHz Pentium III laptop can generate over 10 million random numbers from normal distribution in less than one second. ([http://www.mathworks.com/company/newsletters/news_notes/clevescorner/spring01_cleve.html Reference]) | |||
===Sampling From Binomial Distributions=== | ===Sampling From Binomial Distributions=== | ||
In order to generate a sample x from X ~ Bin(n, p), we can follow the following procedure: | In order to generate a sample x from <math>\displaystyle X \sim Bin(n, p)</math>, we can follow the following procedure: | ||
1. Generate n uniform random numbers sampled from <math>\displaystyle Unif [0, 1] </math>: <math>\displaystyle u_1, u_2, ..., u_n</math>. | |||
2. Set x to be the total number of cases where <math>\displaystyle u_i <= p</math> for all <math>\displaystyle 1 <= i <= n</math>. | |||
In MatLab this can be coded with a single line. The following generates a sample from <math>\displaystyle X \sim Bin(n, p)</math> | |||
sum(rand(n, 1) <= p, 1) | |||
==Bayesian Inference and Frequentist Inference - October 4, 2011== | |||
===Bayesian inference vs Frequentist inference=== | |||
The Bayesian method has become popular in the last few decades as simulation and computer technology makes it more applicable. For more information about its history and application, please refer to http://en.wikipedia.org/wiki/Bayesian_inference. | |||
As for frequentist inference, please refer to http://en.wikipedia.org/wiki/Frequentist_inference. | |||
====Example==== | |||
Consider the 'probability' that a person drinks a cup of coffee on a specific day. The interpretations of this for a frequentist and a bayesian are as follows: | |||
<br><br> | |||
Frequentist: There is no explanation to this expression. It is essentially meaningless since it has only occurred once. Therefore, it is not a probability. | |||
<br> | |||
Bayesian: Probability captures not only the frequency of occurrences but also one's degree of belief about the random component of a proposition. Therefore it is a valid probability. | |||
====Example of face identification==== | |||
Consider a picture of a face that is associated with an identity (person). Take the face as input x and the person as output y. The person can be either Ali or Tom. We have y=1 if it is Ali and y=0 if it is Tom. We can divide the picture into 100*100 pixels and insert them into a 10,000*1 column vector, which captures x. | |||
If you are a frequentist, you would compare <math>\Pr(X=x|y=1)</math> with <math>\Pr(X=x|y=0)</math> and see which one is higher.<br /> If you are Bayesian, you would compare <math>\Pr(y=1|X=x)</math> with <math>\Pr(y=0|X=x)</math>. | |||
====Summary of differences between two schools==== | |||
*Frequentist: Probability refers to limiting relative frequency. (objective) | |||
*Bayesian: Probability describes degree of belief not frequency. (subjective) | |||
e.g. The probability that you drank a cup of tea on May 20, 2001 is 0.62 does not refer to any frequency. | |||
---- | |||
*Frequentist: Parameters are fixed, unknown constants. | |||
*Bayesian: Parameters are random variables and we can make probabilistic statement about them. | |||
---- | |||
*Frequentist: Statistical procedures should be designed to have long run frequency properties. | |||
e.g. a 95% confidence interval should trap true value of the parameter with limiting frequency at least 95%. | |||
*Bayesian: It makes inferences about <math>\theta</math> by producing a probability distribution for <math>\theta</math>. Inference (e.g. point estimates and interval estimates) will be extracted from this distribution :<math>f(\theta|X) = \frac{f(X | \theta)\, f(\theta)}{f(X)}.</math> | |||
====Bayesian inference==== | |||
Bayesian inference is usually carried out in the following way: | |||
1. Choose a prior probability density function of <math>\!\theta</math> which is <math>f(\!\theta)</math>. This is our belief about <math>\theta</math> before we see any data. | |||
2. Choose a statistical model <math>\displaystyle f(x|\theta)</math> that reflects our beliefs about X. | |||
3. After observing data <math>\displaystyle x_1,...,x_n</math>, we update our beliefs and calculate the posterior probability. | |||
<math>f(\theta|x) = \frac{f(\theta,x)}{f(x)}=\frac{f(x|\theta) \cdot f(\theta)}{f(x)}=\frac{f(x|\theta) \cdot f(\theta)}{\int^{}_\theta f(x|\theta) \cdot f(\theta) d\theta}</math>, where <math>\displaystyle f(\theta|x)</math> is the posterior probability, <math>\displaystyle f(\theta)</math> is the prior probability, <math>\displaystyle f(x|\theta)</math> is the likelihood of observing X=x given <math>\!\theta</math> and f(x) is the marginal probability of X=x. | |||
If we have i.i.d. observations <math>\displaystyle x_1,...,x_n</math>, we can replace <math>\displaystyle f(x|\theta)</math> with <math>f({x_1,...,x_n}|\theta)=\prod_{i=1}^n f(x_i|\theta)</math> because of independency. | |||
We denote <math>\displaystyle f({x_1,...,x_n}|\theta)</math> as <math>\displaystyle L_n(\theta)</math> which is called likelihood. And we use <math>\displaystyle x^n</math> to denote <math>\displaystyle (x_1,...,x_n)</math>. | |||
<math>f(\theta|x^n) = \frac{f(x^n|\theta) \cdot f(\theta)}{f(x^n)}=\frac{f(x^n|\theta) \cdot f(\theta)}{\int^{}_\theta f(x^n|\theta) \cdot f(\theta) d\theta}</math> , where <math>\int^{}_\theta f(x^n|\theta) \cdot f(\theta) d\theta</math> is a constant <math>\displaystyle c_n</math> which does not depend on <math>\displaystyle \theta </math>. So <math>f(\theta|x^n) \propto f(x^n|\theta) \cdot f(\theta)</math>. The posterior probability is proportional to the likelihood times prior probability. | |||
Note that it does not matter if we throw away <math>\displaystyle c_n</math>,we can always recover it. | |||
What do we do about the posterier distribution? | |||
*Point estimate | |||
<math> \bar{\theta}=\int\theta \cdot f(\theta|x^n) d\theta=\frac{\int\theta \cdot L_n(\theta)\cdot f(\theta) d(\theta)}{c_n}</math> | |||
*Baysian Interval estimate | |||
<math> \int^{a}_{-\infty} f(\theta|x^n) d\theta=\int^{\infty}_{b} f(\theta|x^n) d\theta=1-\alpha </math> | |||
Let C=(a,b); | |||
Then <math> P(\theta\in C|x^n)=\int^{b}_{a} f(\theta|x^n)d(\theta)=1-\alpha </math>. | |||
C is a <math>\displaystyle 1-\alpha</math> posterior interval. | |||
Let <math>\tilde{\theta}=(\theta_1,...,\theta_d)^T</math>, then <math>f(\theta_1|x^n) = \int^{} \int^{} \dots \int^{}f(\tilde{\theta}|X)d\theta_2d\theta_3 \dots d\theta_d </math> and <math>E(\theta_1|x^n)=\int^{}\theta_1 \cdot f(\theta_1|x^n) d\theta_1</math> | |||
====Example 1: Estimating parameters of a univariate Gaussian distribution==== | |||
Suppose X follows a univariate Gaussian distribution (i.e. a Normal distribution) with parameters <math>\!\mu</math> and | |||
<math>\displaystyle {\sigma^2}</math>. | |||
(a) For Frequentists: | |||
<math>f(x|\theta)= \frac{1}{\sqrt{2\pi}\sigma} \cdot e^{-\frac{1}{2}(\frac{x-\mu}{\sigma})^2}</math> | |||
<math>L_n(\theta)= \prod_{i=1}^n \frac{1}{\sqrt{2\pi}\sigma} \cdot e^{-\frac{1}{2}(\frac{x_i-\mu}{\sigma})^2}</math> | |||
<math>\ln L_n(\theta) = l(\theta) = \sum_{i=1}^n[ -\frac{1}{2}\ln 2\pi-\ln \sigma-\frac{1}{2}(\frac{x_i-\mu}{\sigma})^2]</math> | |||
To get the maximum likelihood estimator of <math>\!\mu</math> (mle), we find the <math>\hat{\mu}</math> which maximizes <math>\displaystyle L_n(\theta)</math>: | |||
<math>\frac{\partial l(\theta)}{\partial \mu}= \sum_{i=1}^n \frac{1}{\sigma}(\frac{x_i-\mu}{\sigma})=0 \Rightarrow \sum_{i=1}^n x_i = n\mu \Rightarrow \hat{\mu}_{mle}=\bar{x}</math> | |||
(b) For Bayesians: | |||
<math>f(\theta|x) \propto f(x|\theta) \cdot f(\theta)</math> | |||
We assume that the mean of the above normal distribution is itself distributed normally with mean <math>\!\mu_0</math> and variance <math>\!\Gamma</math>. | |||
Suppose <math>\!\mu\sim N(\mu_0, \!\Gamma^2</math>), | |||
so <math>f(\mu) = \frac{1}{\sqrt{2\pi}\Gamma} \cdot e^{-\frac{1}{2}(\frac{\mu-\mu_0}{\Gamma})^2}</math> | |||
<math>f(x|\mu) = \frac{1}{\sqrt{2\pi}\tilde{\sigma}} \cdot e^{-\frac{1}{2}(\frac{x-\mu}{\tilde{\sigma}})^2}</math> | |||
<math>\tilde{\mu} = \frac{\frac{n}{\sigma^2}}{\frac{n}{\sigma^2}+\frac{1}{\Gamma^2}}\bar{x}+\frac{\frac{1}{\Gamma^2}}{\frac{n}{\sigma^2}+\frac{1}{\Gamma^2}}\mu_0</math>, where <math>\tilde{\mu}</math> is the estimator of <math>\!\mu</math>. | |||
* If prior belief about <math>\!\mu_0</math> is strong, then <math>\!\Gamma</math> is small and <math>\frac{1}{\Gamma^2}</math> is large. <math>\tilde{\mu}</math> is close to <math>\!\mu_0</math> and the observations will not affect too much. On the contrary, if prior belief about <math>\!\mu_0</math> is weak, <math>\!\Gamma</math> is large and <math>\frac{1}{\Gamma^2}</math> is small. <math>\tilde{\mu}</math> depends more on observations. (This is intuitive, when our original belief is reliable, then the sample is not important in improving the result; when the belief is not reliable, then we depend a lot on the sample.) | |||
* When the sample is large (i.e. n <math>\to \infty</math>), <math>\tilde{\mu} \to \bar{x}</math> and the impact of prior belief about <math>\!\mu</math> is weakened. | |||
=='''Basic Monte Carlo Integration - October 6th, 2011'''== | |||
Three integration methods would be taught in this course: | |||
*Basic Monte Carlo Integration | |||
*Importance Sampling | |||
*Markov Chain Monte Carlo (MCMC) | |||
The first, and most basic, method of numerical integration we will see is Monte Carlo Integration. We use this to solve an integral of the form: <math> I = \int_{a}^{b} h(x) dx </math> | |||
Note the following derivation: | |||
<math>\begin{align} | |||
\displaystyle I & = \int_{a}^{b} h(x)dx \\ | |||
& = \int_{a}^{b} h(x)((b-a)/(b-a))dx \\ | |||
& = \int_{a}^{b} (h(x)(b-a))(1/(b-a))dx \\ | |||
& = \int_{a}^{b} w(x)f(x)dx \\ | |||
& = E[w(x)] \\ | |||
\end{align} | |||
</math> | |||
~ <math> \frac{1}{n} \sum_{i=1}^{n} w(x) </math> | |||
Where w(x) = h(x)(b-a) and f(x) is the probability density function of a uniform random variable on the interval [a,b]. The expectation, with respect to the distribution of f, of w is taken from n samples of x. | |||
===='''General Procedure'''==== | |||
i) Draw n samples <math> x_i \sim~ Unif[a,b] </math> | |||
ii) Compute <math> \ w(x_i) </math> for every sample | |||
iii) Obtain an estimate of the integral, <math> \hat{I} </math>, as follows: | |||
<math> \hat{I} = \frac{1}{n} \sum_{i=1}^{n} w(x_i)</math> . Clearly, this is just the average of the simulation results. | |||
By the strong law of large numbers <math> \hat{I} </math> converges to <math> \ I </math> as <math> \ n \rightarrow \infty </math>. Because of this, we can compute all sorts of useful information, such as variance, standard error, and confidence intervals. | |||
Standard Error: <math> SE = \frac{Standard Deviation} {\sqrt{n}} </math> | |||
Variance: <math> V = \frac{\sum_{i=1}^{n} (w(x)-\hat{I})^2}{n-1} </math> | |||
Confidence Interval: <math> \hat{I} \pm t_{(\alpha/2)} SE </math> | |||
==='''Example: Uniform Distribution'''=== | |||
Consider the integral, <math> \int_{0}^{1} x^3dx </math>, which is easily solved through standard analytical integration methods, and is equal to .25. Now, let us check this answer with a numerical approximation using Monte Carlo Integration. | |||
We generate a 1 by 10000 vector of uniform (on the interval [0,1]) random variables and call that vector 'u'. We see that our 'w' in this case is <math> x^3 </math>, so we set <math> w = u^3 </math>. Our <math>\hat{I}</math> is equal to the mean of w. | |||
In Matlab, we can solve this integration problem with the following code: | |||
<pre> | |||
u = rand(1,10000); | |||
w = u.^3; | |||
mean(w) | |||
ans = 0.2475 | |||
</pre> | |||
Note the '.' after 'u' in the second line of code, indicating that each entry in the matrix is cubed. Also, our approximation is close to the actual value of .25. Now let's try to get an even better approximation by generating more sample points. | |||
<pre> | |||
u= rand(1,100000); | |||
w= u.^3; | |||
mean(w) | |||
ans = .2503 | |||
</pre> | |||
We see that when the number of sample points is increased, our approximation improves, as one would expect. | |||
==='''Generalization'''=== | |||
Up to this point we have seen how to numerically approximate an integral when the distribution of f is uniform. Now we will see how to generalize this to other distributions. | |||
<math> I = \int h(x)f(x)dx </math> | |||
If f is a distribution function (pdf), then <math> I </math> can be estimated as E<sub>f</sub>[h(x)]. This means taking the expectation of h with respect to the distribution of f. Our previous example is the case where f is the uniform distribution between [a,b]. | |||
'''Procedure for the General Case''' | |||
i) Draw n samples from f | |||
ii) Compute h(x<sub>i</sub>) | |||
iii) <math>\hat{I} = \frac{1}{n} \sum_{i=1}^{n} h(x</math><sub>i</sub><math>)</math> | |||
==='''Example: Exponential Distribution'''=== | |||
Find <math> E[\sqrt{x}] </math> for <math> \displaystyle f = e^{-x} </math>, which is the exponential distribution with mean 1. | |||
<math> I = \int_{0}^{\infty} \sqrt{x} e^{-x}dx </math> | |||
We can see that we must draw samples from f, the exponential distribution. | |||
To find a numerical solution using Monte Carlo Integration we see that: | |||
u= rand(1,10000)<br /> | |||
X= -log(u)<br /> | |||
h= <math> \sqrt{x} </math> <br /> | |||
I= mean(h) | |||
To implement this procedure in Matlab, use the following code: | |||
<pre> | |||
u = rand(1,10000); | |||
X = -log(u); | |||
h = x.^.5; | |||
mean(h) | |||
ans = .8841 | |||
</pre> | |||
An easy way to check whether your approximation is correct is to use the built in Matlab function 'quadl' which takes a function and bounds for the integral and returns a solution for the definite integral of that function. For this specific example, we can enter: | |||
<pre> | |||
f = @(x) sqrt(x).*exp(-x); | |||
% quadl runs into computational problems when the upper bound is "inf" or an extremely large number, | |||
% so choose just a moderately large number. | |||
quadl(f,0,100) | |||
ans = | |||
0.8862 | |||
</pre> | |||
From the above result, we see that our approximation was quite close. | |||
==='''Example: Normal Distribution'''=== | |||
Let <math> f(x) = (1/(2 \pi)^{1/2}) e^{(-x^2)/2} </math>. Compute the cumulative distribution function at some point x. | |||
<math> F(x)= \int_{-\infty}^{x} f(s)ds = \int_{-\infty}^{x}(1)(1/(2 \pi)^{1/2}) e^{(-s^2)/2}ds </math>. The (1) is inserted to illustrate that our h(x) will be the constant function 1, and our f(x) is the normal distribution. To take into account the upper bound of integration, x, any values sampled that are greater than x will be set to zero. | |||
This is the Matlab code for solving F(2): | |||
<pre> | |||
u = randn(1,10000) | |||
h = u < 2; | |||
mean(h) | |||
ans = .9756 | |||
</pre> | |||
We generate a 1 by 10000 vector of standard normal random variables and we return a value of 1 if u is less than 2, and 0 otherwise. | |||
We can also build the function F(x) in matlab in the following way: | |||
<pre> | |||
function F(x) | |||
u=rand(1,1000000); | |||
h=u<x; | |||
mean(h) | |||
</pre> | |||
==='''Example: Binomial Distribution'''=== | |||
In this example we will see the Bayesian Inference for 2 Binomial Distributions. | |||
Let <math> X ~ Bin(n,p) </math> and <math> Y ~ Bin(m,q) </math>, and let <math> \!\delta = p-q </math>. | |||
Therefore, <math> \displaystyle \!\delta = x/n - y/m </math> which is the frequentist approach. | |||
Bayesian wants <math> \displaystyle f(p,q|x,y) = f(x,y|p,q)f(p,q)/f(x,y) </math>, where <math> f(x,y)=\iint\limits_{\!\theta} f(x,y|p,q)f(p,q)\,dp\,dq</math> is a constant. | |||
Thus, <math> \displaystyle f(p,q|x,y)\propto f(x,y|p,q)f(p,q) </math>. Now we assume that <math>\displaystyle f(p,q) = f(p)f(q) = 1 </math> and f(p) and f(q) are uniform. | |||
Therefore, <math> \displaystyle f(p,q|x,y)\propto p^x(1-p)^{n-x}q^y(1-q)^{m-y} </math>. | |||
<math> E[\delta|x,y] = \int_{0}^{1} \int_{0}^{1} (p-q)f(p,q|x,y)dpdq </math>. | |||
As you can see this is much tougher than the frequentist approach. | |||
=='''Importance Sampling and Basic Monte Carlo Integration - October 11th, 2011'''== | |||
==='''Example: Binomial Distribution (Continued)'''=== | |||
Suppose we are given two independent Binomial Distributions <math>\displaystyle X \sim Bin(n, p_1)</math>, <math>\displaystyle Y \sim Bin(m, p_2)</math>. We would like to give an Monte Carlo estimate of <math>\displaystyle \delta = p_1 - p_2</math><br> | |||
Frequentist approach: <br><br><math>\displaystyle \hat{p_1} = \frac{X}{n}</math> ; <math>\displaystyle \hat{p_2} = \frac{Y}{m}</math><br><br><math>\displaystyle \hat{\delta} = \hat{p_1} - \hat{p_2} = \frac{X}{n} - \frac{Y}{m}</math><br><br> | |||
Bayesian approach to compute the expected value of <math>\displaystyle \delta</math>:<br><br> | |||
<math>\displaystyle E(\delta|X,Y) = \int\int(p_1-p_2) f(p_1,p_2|X,Y)\,dp_1dp_2</math><br><br> | |||
Assume that <math>\displaystyle n = 100, m = 100, p_1 = 0.5, p_2 = 0.8</math> and the sample size is 1000.<br> | |||
MATLAB code of the above example: | |||
<pre> | |||
n = 100; | |||
m = 100; | |||
p_1 = 0.5; | |||
p_2 = 0.8; | |||
p1 = mean(rand(n,1000)<p_1,1); | |||
p2 = mean(rand(m,1000)<p_2,1); | |||
delta = p2 - p1; | |||
hist(delta) | |||
mean(delta) | |||
</pre> | |||
In one execution of the code, the mean of delta was 0.3017. The histogram of delta generated was: | |||
[[File:Hist delta.jpg|center|]] | |||
Through Monte Carlo simulation, we can obtain an empirical distribution of delta and carry out inference on the data obtained, such as computing the mean, maximum, variance, standard deviation and the standard error of delta. | |||
==='''Importance Sampling'''=== | |||
====Motivation==== | |||
Consider the integral <math>\displaystyle I = \int h(x)f(x)\,dx</math><br><br> | |||
According to basic Monte Carlo Integration, if we can sample from the probability density function <math>\displaystyle f(x)</math> and feed the samples of <math>\displaystyle f(x)</math> back to <math>\displaystyle h(x)</math>, <math>\displaystyle I</math> can be estimated as an average of <math>\displaystyle h(x)</math> ( i.e. <math>\hat{I} = \frac{1}{n} \sum_{i=1}^{n} h(x_i)</math> )<br> | |||
However, the Monte Carlo method works when we know how to sample from <math>\displaystyle f(x)</math>. In the case where it is difficult to sample from <math>\displaystyle f(x)</math>, importance sampling is a technique that we can apply. Importance Sampling relies on another function <math>\displaystyle g(x)</math> which we know how to sample from. | |||
The above integral can be rewritten as follow:<br> | |||
<math>\begin{align} | |||
\displaystyle I & = \int h(x)f(x)\,dx \\ | |||
& = \int h(x)f(x)\frac{g(x)}{g(x)}\,dx \\ | |||
& = \int \frac{h(x)f(x)}{g(x)}g(x)\,dx \\ | |||
& = \int y(x)g(x)\,dx \\ | |||
& = E_g(y(x)) \\ | |||
\end{align} | |||
</math><br> | |||
<math>where \ y(x) = \frac{h(x)f(x)}{g(x)}</math><br> | |||
The integral can thus be simulated as <math>\displaystyle \hat{I} = \frac{1}{n} \sum_{i=1}^{n} Y_i \ , \ where \ Y_i = \frac{h(x_i)f(x_i)}{g(x_i)}</math><br> | |||
====Procedure==== | |||
Suppose we know how to sample from <math>\displaystyle g(x)</math><br> | |||
#Choose a suitable <math>\displaystyle g(x)</math> and draw n samples <math>x_1,x_2....,x_n \sim~ g(x)</math> | |||
#Set <math>Y_i =\frac{h(x_i)f(x_i)}{g(x_i)}</math> | |||
#Compute <math> \hat{I} = \frac{1}{n}\sum_{i=1}^{n} Y_i </math><br> | |||
By the Law of large numbers, <math>\displaystyle \hat{I} \rightarrow I </math> provided that the sample size n is large enough.<br><br> | |||
'''Remarks:''' One can think of <math>\frac{f(x)}{g(x)}</math> as a weight to <math>\displaystyle h(x)</math> in the computation of <math>\hat{I}</math><br><br> | |||
<math>\displaystyle i.e. \ \hat{I} = \frac{1}{n}\sum_{i=1}^{n} Y_i = \frac{1}{n}\sum_{i=1}^{n} (\frac{f(x_i)}{g(x_i)})h(x_i)</math><br><br> | |||
Therefore, <math>\displaystyle \hat{I} </math> is a weighted average of <math>\displaystyle h(x_i)</math><br><br> | |||
====Problem==== | |||
If <math>\displaystyle g(x)</math> is not chosen appropriately, then the variance of the estimate <math>\hat{I}</math> may be very large. The problem here is actually similar to what we encounter with the Rejection-Acceptance method. Consider the second moment of <math>y(x)</math>:<br><br> | |||
<math>\begin{align} | |||
E_g((y(x))^2) \\ | |||
& = \int (\frac{h(x)f(x)}{g(x)})^2 g(x) dx \\ | |||
& = \int \frac{h^2(x) f^2(x)}{g^2(x)} g(x) dx \\ | |||
& = \int \frac{h^2(x)f^2(x)}{g(x)} dx \\ | |||
\end{align} | |||
</math><br><br> | |||
When <math>\displaystyle g(x)</math> is very small, then the above integral could be very large, hence the variance can be very large when g is not chosen appropriately. This occurs when <math>\displaystyle g(x)</math> has a thinner tail than <math>\displaystyle f(x)</math> such that the quantity <math>\displaystyle \frac{h^2(x)f^2(x)}{g(x)}</math> is large. | |||
'''Remarks:''' | |||
1. We can actually compute the form of <math>\displaystyle g(x)</math> to have optimal variance. <br>Mathematically, it is to find <math>\displaystyle g(x)</math> subject to <math>\displaystyle \min_g [\ E_g([y(x)]^2) - (E_g[y(x)])^2\ ]</math><br> | |||
It can be shown that the optimal <math>\displaystyle g(x)</math> is <math>\displaystyle {|h(x)|f(x)}</math>. Using the optimal <math>\displaystyle g(x)</math> will minimize the variance of estimation in Importance Sampling. This is of theoretical interest but not useful in practice. As we can see, if we can actually show the expression of g(x), we must first have the value of the integration---which is what we want in the first place. | |||
2. In practice, we shall choose <math>\displaystyle g(x)</math> which has similar shape as <math>\displaystyle f(x)</math> but with a thicker tail than <math>\displaystyle f(x)</math> in order to avoid the problem mentioned above.<br> | |||
====Example==== | |||
Estimate <math>\displaystyle I = Pr(Z>3),\ where\ Z \sim N(0,1)</math><br><br> | |||
'''Method 1: Basic Monte Carlo''' | |||
<math>\begin{align} Pr(Z>3) & = \int^\infty_3 f(x)\,dx \\ | |||
& = \int^\infty_{-\infty} h(x)f(x)\,dx \end{align}</math><br /> | |||
<math> where \ | |||
h(x) = \begin{cases} | |||
0, & \text{if } x \le 3 \\ | |||
1, & \text{if } x > 3 | |||
\end{cases}</math> | |||
<math>\ ,\ f(x) = \frac{1}{\sqrt{2\pi}}e^{-\frac{1}{2}x^2}</math> | |||
MATLAB code to compute <math>\displaystyle I</math> from 100 samples of standard normal distribution: | |||
<pre> | |||
h = randn(100,1) > 3; | |||
I = mean(h) | |||
</pre> | |||
In one execution of the code, it returns a value of 0 for <math>\displaystyle I</math>, which differs significantly from the true value of <math>\displaystyle I \approx 0.0013 </math>. The problem of using Basic Monte Carlo in this example is that <math>\displaystyle Pr(Z>3)</math> has a small value, and hence many points sampled from the standard normal distribution will be wasted. Therefore, although Basic Monte Carlo is a feasible method to compute <math>\displaystyle I</math>, it gives a poor estimation. | |||
'''Method 2: Importance Sampling''' | |||
<math>\displaystyle I = Pr(Z>3)= \int^\infty_3 f(x)\,dx </math><br> | |||
To apply importance sampling, we have to choose a <math>\displaystyle g(x)</math> which we will sample from. In this example, we can choose <math>\displaystyle g(x)</math> to be the probability density function of exponential distribution, normal distribution with mean 0 and variance greater than 1 or normal distribution with mean greater than 0 and variance 1 etc. The goal is to minimize the number of rejected samples in order to produce a more accurate result! For the following, we take <math>\displaystyle g(x)</math> to be the pdf of <math>\displaystyle N(4,1)</math>.<br> | |||
Procedure: | |||
#Draw n samples <math>x_1,x_2....,x_n \sim~ g(x)</math> | |||
#Calculate <math>\begin{align} \frac{f(x)}{g(x)} & = \frac{ \frac{1}{\sqrt{2\pi}}e^{-\frac{1}{2}x^2} | |||
}{ \frac{1}{\sqrt{2\pi}}e^{-\frac{1}{2}(x-4)^2} } \\ | |||
& = e^{8-4x} \end{align} </math><br> | |||
#Set <math> Y_i = h(x_i)e^{8-4x_i}\ with\ h(x) = \begin{cases} | |||
0, & \text{if } x \le 3 \\ | |||
1, & \text{if } x > 3 | |||
\end{cases} | |||
</math><br> | |||
#Compute <math> \hat{Y} = \frac{1}{n}\sum_{i=1}^{n} Y_i </math><br> | |||
The above procedure from 100 samples of <math>\displaystyle g(x)</math>can be implemented in MATLAB as follow: | |||
<pre> | |||
for ii = 1:100 | |||
x = randn + 4 ; | |||
h = x > 3 ; | |||
y(ii) = h * exp(8-4*x) ; | |||
end | |||
mean(y) | |||
</pre> | |||
In one execution of the code, it returns a value of 0.001271 for <math> \hat{Y} </math>, which is much closer to the true value of <math>\displaystyle I \approx 0.0013 </math>. From many executions of the code, the variance of basic monte carlo is approximately 150 times that of importance sampling. This demonstrates that this method can provide a better estimate than the Basic Monte Carlo method. | |||
==''' Importance Sampling with Normalized Weight and Markov Chain Monte Carlo - October 13th, 2011'''== | |||
==='''Importance Sampling with Normalized Weight'''=== | |||
Recall that we can think of <math>\displaystyle b(x) = \frac{f(x)}{g(x)}</math> as a weight applied to the samples <math>\displaystyle h(x)</math>. If the form of <math>\displaystyle f(x)</math> is known only up to a constant, we can use an alternate, normalized form of the weight, <math>\displaystyle b^*(x)</math>. (This situation arises in Bayesian inference.) Importance sampling with normalized or standard weight is also called indirect importance sampling. | |||
We derive the normalized weight as follows:<br> | |||
<math>\begin{align} | |||
\displaystyle I & = \int h(x)f(x)\,dx \\ | |||
&= \int h(x)\frac{f(x)}{g(x)}g(x)\,dx \\ | |||
&= \frac{\int h(x)\frac{f(x)}{g(x)}g(x)\,dx}{\int f(x) dx} \\ | |||
&= \frac{\int h(x)\frac{f(x)}{g(x)}g(x)\,dx}{\int \frac{f(x)}{g(x)}g(x) dx} \\ | |||
&= \frac{\int h(x)b(x)g(x)\,dx}{\int\ b(x)g(x) dx} | |||
\end{align}</math> | |||
<math>\hat{I}= \frac{\sum_{i=1}^{n} h(x_i)b(x_i)}{\sum_{i=1}^{n} b(x_i)} </math> | |||
Then, the normalized weight is <math>b^*(x) = \displaystyle \frac{b(x_i)}{\sum_{i=1}^{n} b(x_i)}</math> | |||
Note that <math> \int f(x) dx = 1 = \int b(x)g(x) dx = 1 </math> | |||
We can also determine the associated Monte Carlo variance of this estimate by | |||
<math> Var(\hat{I})= \frac{\sum_{i=1}^{n} b(x_i)(h(x_i) - \hat{I})^2}{\sum_{i=1}^{n} b(x_i)} </math> | |||
==='''Markov Chain Monte Carlo'''=== | |||
We still want to solve <math> I = \displaystyle\int^\ h(x)f(x)\,dx </math> | |||
====Stochastic Process==== | |||
A stochastic process <math> \{ x_t : t \in T \}</math> is a collection of random variables. Variables <math>\displaystyle x_t</math> take values in some set <math>\displaystyle X</math> called the '''state space.''' The set <math>\displaystyle T</math> is called the '''index set.''' | |||
===='''Markov Chain'''==== | |||
A Markov Chain is a stochastic process for which the distribution of <math>\displaystyle x_t</math> depends only on <math>\displaystyle x_{t-1}</math>. It is a random process characterized as being memoryless, meaning that the next occurrence of a defined event is only dependent on the current event;not on the preceding sequence of events. | |||
Formal Definition: The process <math> \{ x_t : t \in T \}</math> is a Markov Chain if <math>\displaystyle Pr(x_t|x_0, x_1,..., x_{t-1})= Pr(x_t|x_{t-1})</math> for all <math> \{t \in T \}</math> and for all <math> \{x \in X \}</math> | |||
For a Markov Chain, <math>\displaystyle f(x_1,...x_n)= f(x_1)f(x_2|x_1)f(x_3|x_2)...f(x_n|x_{n-1})</math> | |||
<br><br>'''Real Life Example:''' | |||
<br>When going for an interview, the employer only looks at your highest education achieved. The employer would not look at the past educations received (elementary school, high school etc.) because the employer believes that the highest education achieved summarizes your previous educations. Therefore, anything before your most recent previous education is irrelevant. In other word, we can say that <math>\! x_t </math> is regarded as the summary of <math>\!x_{t-1},...,x_2,x_1</math>, so when we need to determine <math>\!x_{t+1}</math>, we only need to pay attention in <math>\!x_{t}</math>. | |||
====Transition Probabilities==== | |||
The <b>transition probability</b> is the probability of jumping from one state to another state in a Markov Chain. | |||
Formally, let us define <math>\displaystyle P_{ij} = Pr(x_{t+1}=j|x_t=i)</math> to be the transition probability. | |||
That is, <math>\displaystyle P_{ij}</math> is the probability of transitioning from state i to state j in a single step. Then the matrix <math>\displaystyle P</math> whose (i,j) element is <math>\displaystyle P_{ij}</math> is called the <b>transition matrix</b>. | |||
Properties of P: | |||
:1) <math>\displaystyle P_{ij} >= 0</math> (The probability of going to another state cannot be negative) | |||
:2) <math>\displaystyle \sum_{\forall j}P_{ij} = 1</math> (The probability of transitioning to some state from state i (including remaining in state i) is one) | |||
====Random Walk==== | |||
Example: Start at one point and flip a coin where <math>\displaystyle Pr(H)=p</math> and <math>\displaystyle Pr(T)=1-p=q</math>. Take one step right if heads and one step left if tails. If at an endpoint, stay there. | |||
The transition matrix is | |||
<math>P=\left(\begin{matrix}1&0&0&\dots&\dots&0\\ | |||
q&0&p&0&\dots&0\\ | |||
0&q&0&p&\dots&0\\ | |||
\vdots&\vdots&\vdots&\ddots&\vdots&\vdots\\ | |||
\vdots&\vdots&\vdots&\vdots&\ddots&\vdots\\ | |||
0&0&\dots&\dots&\dots&1 | |||
\end{matrix}\right)</math> | |||
Let <math>\displaystyle P_n</math> be the matrix such that its (i,j) element is <math>\displaystyle P_{ij}(n)</math>. This is called n-step probability. | |||
:<math>\displaystyle P_n = P^n</math> | |||
:<math>\displaystyle P_1 = P</math> | |||
:<math>\displaystyle P_2 = P^2</math> | |||
==''' Markov Chain Properties and Page Rank - October 18th, 2011'''== | |||
===Summary of Terminology=== | |||
====Transition Matrix==== | |||
A matrix <math>\!P</math> that defines a Markov Chain has the form: | |||
<math>P = \begin{bmatrix} | |||
P_{11} & \cdots & P_{1N} \\ | |||
\vdots & \ddots & \vdots \\ | |||
P_{N1} & \cdots & P_{NN} | |||
\end{bmatrix}</math> | |||
where <math>\!P(i,j) = P_{ij} = Pr(x_{t+1} = j | x_t = i) </math> is the probability of transitioning from state i to state j in the Markov Chain in a single step. Note that this implies that all rows add up to one. | |||
====n-step Transition matrix==== | |||
A matrix <math>\!P_n</math> whose (i,j)<sup>th</sup> entry is the probability of moving from state i to state j after n transitions: | |||
<math>\!P_n(i,j) = Pr(x_{m+n}=j|x_m = i)</math> | |||
This probability is called the n-step transition probability. A nice property of this matrix is that | |||
<math>\!P_n = P^n</math> | |||
For all n >= 0, where P is the transition matrix. Note that the rows of <math>\!P_n</math> should still add up to one. | |||
====Marginal distribution of a Markov Chain==== | |||
We represent the state at time t as a vector. | |||
<math>\mu_t = (\mu_t(1) \; \mu_t(2) \; ... \; \mu_t(n))</math> | |||
Consider this Markov Chain: | |||
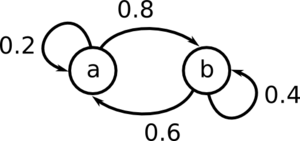
[[File:MarkovSample.png|300px]] | |||
<math>\mu_t = (A \; B)</math>, where A is the probability of being in state a at time t, and B is the probability of being in state b at time t. | |||
For example if <math>\mu_t = (0.1 \; 0.9)</math>, we have a 10% chance of being in state a at time t, and a 90% chance of being in state b at time t. | |||
Suppose we run this Markov chain many times, and record the state at each step. | |||
In this example, we run 4 trials, up until t=5. | |||
{| class="wikitable" | |||
|- | |||
! t | |||
! Trial 1 | |||
! Trial 2 | |||
! Trial 3 | |||
! Trial 4 | |||
! Observed <math>\mu</math> | |||
|- | |||
| 1 | |||
| a | |||
| b | |||
| b | |||
| a | |||
| (0.5, 0.5) | |||
|- | |||
| 2 | |||
| b | |||
| a | |||
| a | |||
| a | |||
| (0.75, 0.25) | |||
|- | |||
| 3 | |||
| a | |||
| a | |||
| b | |||
| a | |||
| (0.75, 0.25) | |||
|- | |||
| 4 | |||
| b | |||
| b | |||
| a | |||
| b | |||
| (0.25, 0.75) | |||
|- | |||
| 5 | |||
| b | |||
| b | |||
| b | |||
| a | |||
| (0.25, 0.75) | |||
|} | |||
Imagine simulating the chain many times. If we collect all the outcomes at time t from all the chains, the histogram of this data would look like <math>\!\mu_t</math>. | |||
We can find the marginal probabilities as <math>\!\mu_n = \mu_0 P^n</math> | |||
====Stationary Distribution==== | |||
Let <math>\pi = (\pi_i \mid i \in \chi)</math> be a vector of non-negative numbers that sum to 1. (i.e. <math>\!\pi</math> is a pmf) | |||
If <math>\!\pi = \pi P</math>, then <math>\!\pi</math> is a stationary distribution, also known as an invariant distribution. | |||
====Limiting Distribution==== | |||
A Markov chain has limiting distribution <math>\!\pi </math> if <math>\lim_{n \to \infty} P^n = \begin{bmatrix} \pi \\ \vdots \\ \pi \end{bmatrix}</math> | |||
That is, <math>\!\pi_j = \lim_{n \to \infty}\left [ P^n \right ]_{ij}</math> exists and is independent of i. | |||
Here is an example: | |||
Suppose we want to find the stationary distribution of | |||
<math>P=\left(\begin{matrix} | |||
1/3&1/3&1/3\\ | |||
1/4&3/4&0\\ | |||
1/2&0&1/2 | |||
\end{matrix}\right)</math> | |||
We want to solve <math>\pi=\pi P</math> and we want <math>\displaystyle \pi_0 + \pi_1 + \pi_2 = 1</math><br /> | |||
<math>\displaystyle \pi_0 = \frac{1}{3}\pi_0 + \frac{1}{4}\pi_1 + \frac{1}{2}\pi_2</math><br /> | |||
<math>\displaystyle \pi_1 = \frac{1}{3}\pi_0 + \frac{3}{4}\pi_1</math><br /> | |||
<math>\displaystyle \pi_2 = \frac{1}{3}\pi_0 + \frac{1}{2}\pi_2</math><br /> | |||
Solving the system of equations, we get <br /> | |||
<math>\displaystyle \pi_1 = \frac{4}{3}\pi_0</math><br /> | |||
<math>\displaystyle \pi_2 = \frac{2}{3}\pi_0</math><br /> | |||
So using our condition above, we have <math>\displaystyle \pi_0 + \frac{4}{3}\pi_0 + \frac{2}{3}\pi_0 = 1</math> and by solving we get <math>\displaystyle \pi_0 = \frac{1}{3}</math> | |||
Using this in our system of equations, we obtain: <br /> | |||
<math>\displaystyle \pi_1 = \frac{4}{9}</math><br /> | |||
<math>\displaystyle \pi_2 = \frac{2}{9}</math> | |||
Thus, the stationary distribution is <math>\displaystyle \pi = (\frac{1}{3}, \frac{4}{9}, \frac{2}{9})</math> | |||
====Detailed Balance==== | |||
<math>\!\pi</math> has the detailed balance property if <math>\!\pi_iP_{ij} = P_{ji}\pi_j</math> | |||
'''Theorem''' | |||
If <math>\!\pi</math> satisfies detailed balance, then <math>\!\pi</math> is a stationary distribution. | |||
In other words, if <math>\!\pi_iP_{ij} = P_{ji}\pi_j</math>, then <math>\!\pi = \pi P</math> | |||
'''Proof:''' | |||
<math>\!\pi P = | |||
\begin{bmatrix}\pi_1 & \pi_2 & \cdots & \pi_N\end{bmatrix} \begin{bmatrix}P_{11} & \cdots & P_{1N} \\ \vdots & \ddots & \vdots \\ P_{N1} & \cdots & P_{NN}\end{bmatrix}</math> | |||
Observe that the j<sup>th</sup> element of <math>\!\pi P</math> is | |||
<math>\!\left [ \pi P \right ]_j = \pi_1 P_{1j} + \pi_2 P_{2j} + \dots + \pi_N P_{Nj}</math> | |||
::<math>\! = \sum_{i=1}^N \pi_i P_{ij}</math> | |||
::<math>\! = \sum_{i=1}^N P_{ji} \pi_j</math>, by the definition of detailed balance. | |||
::<math>\! = \pi_j \sum_{i=1}^N P_{ji}</math> | |||
::<math>\! = \pi_j</math>, as the sum of the entries in a row of P must sum to 1. | |||
So <math>\!\pi = \pi P</math>. | |||
'''Example''' | |||
Find the marginal distribution of | |||
[[File:MarkovSample.png|300px]] | |||
Start by generating the matrix P. | |||
<math>\!P = \begin{pmatrix} 0.2 & 0.8 \\ 0.6 & 0.4 \end{pmatrix}</math> | |||
We must assume some starting value for <math>\mu_0</math> | |||
<math>\!\mu_0 = \begin{pmatrix} 0.1 & 0.9 \end{pmatrix}</math> | |||
For t = 1, the marginal distribution is | |||
<math>\!\mu_1 = \mu_0 P</math> | |||
Notice that this <math>\mu</math> converges. | |||
If you repeatedly run: | |||
<math>\!\mu_{i+1} = \mu_i P</math> | |||
It converges to <math>\mu = \begin{pmatrix} 0.4286 & 0.5714 \end{pmatrix}</math> | |||
This can be seen by running the following Matlab code: | |||
P = [0.2 0.8; 0.6 0.4]; | |||
mu = [0.1 0.9]; | |||
while 1 | |||
mu_old = mu; | |||
mu = mu * P; | |||
if mu_old == mu | |||
disp(mu); | |||
break; | |||
end | |||
end | |||
Another way of looking at this simple question is that we can see whether the ultimate pmf converges: | |||
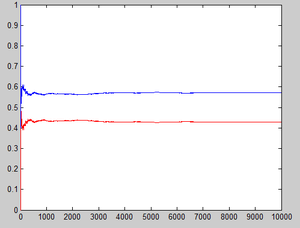
Let <math>\hat{p_n}(1)=\frac{1}{n}\sum_{k=1}^n I(X_k=1)</math> denote the estimator of the stationary probability of state 1,<math>\hat{p_n}(2)=\frac{1}{n}\sum_{k=1}^n I(X_k=2)</math> denote the estimator of the stationary probability of state 2, where <math>\displaystyle I(X_k=1)</math> and <math>\displaystyle I(X_k=2)</math> are indicator variables which equal 1 if <math>X_k=1</math>(or <math>X_k=2</math> for the latter one). | |||
Matlab codes for this explanation is | |||
n=1; | |||
if rand<0.1 | |||
x(1)=1; | |||
else | |||
x(1)=0; | |||
end | |||
p1(1)=sum(x)/n; | |||
p2(1)=1-p1(1); | |||
for i=2:10000 | |||
n=n+1; | |||
if (x(i-1)==1&rand<0.2)|(x(i-1)==0&rand<0.6) | |||
x(i)=1; | |||
else | |||
x(i)=0; | |||
end | |||
p1(i)=sum(x)/n; | |||
p2(i)=1-p1(i); | |||
end | |||
plot(p1,'red'); | |||
hold on; | |||
plot(p2) | |||
The results can be easily seen from the graph below: | |||
[[File:Stationary distribution.png|300px]] | |||
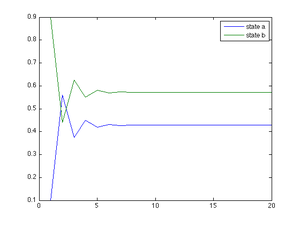
Additionally, we can plot the marginal distribution as it converges without estimating it. The following Matlab code shows this: | |||
%transition matrix | |||
P=[0.2 0.8; 0.6 0.4]; | |||
%mu at time 0 | |||
mu=[0.1 0.9]; | |||
%number of points for simulation | |||
n=20; | |||
for i=1:n | |||
mu_a(i)=mu(1); | |||
mu_b(i)=mu(2); | |||
mu=mu*P; | |||
end | |||
t=[1:n]; | |||
plot(t, mu_a, t, mu_b); | |||
hleg1=legend('state a', 'state b'); | |||
[[File:Marginal distribution convergence.png|300px]] | |||
Note that there are chains with stationary distributions that don't converge (the chain might not naturally reach the stationary distribution, and isn't limiting). An example of this is: | |||
<math>P = \begin{pmatrix} 0 & 1 & 0 \\ 0 & 0 & 1 \\ 1 & 0 & 0 \end{pmatrix}, \mu_0 = \begin{pmatrix} 1/3 & 1/3 & 1/3 \end{pmatrix}</math> | |||
<math>\!\mu_0</math> is a stationary distribution, so <math>\!\mu P</math> is the same for all iterations. | |||
But, | |||
<math>P^{1000} = \begin{pmatrix} 0 & 0 & 1 \\ 1 & 0 & 0 \\ 0 & 1 & 0 \end{pmatrix} \ne \begin{pmatrix} \mu \\ \mu \\ \mu \end{pmatrix}</math> | |||
So <math>\!\mu</math> is not a limiting distribution. Also, if | |||
<math>\mu = \begin{pmatrix} 0.2 & 0.1 & 0.7 \end{pmatrix}</math> | |||
Then <math>\!\mu \neq \mu P</math>. | |||
This can be observed through the following Matlab code. | |||
P = [0 0 1; 1 0 0; 0 1 0]; | |||
mu = [0.2 0.1 0.7]; | |||
for i= 1:4 | |||
mu = mu * P; | |||
disp(mu); | |||
end | |||
Output: | |||
0.1000 0.7000 0.2000 | |||
0.7000 0.2000 0.1000 | |||
0.2000 0.1000 0.7000 | |||
0.1000 0.7000 0.2000 | |||
Note that <math>\!\mu_1 = \!\mu_4</math>, which indicates that <math>\!\mu</math> will cycle forever. | |||
This means that this chain has a stationary distribution, but is not limiting. A chain has a limiting distribution iff it is ergodic, that is, aperiodic and positive recurrent. While cycling breaks detailed balance and limiting distribution on the whole state space does not exist, the cycling behavior itself is the "limiting distribution". Also, for each cycles (closed class), we will have a mini-limiting distribution which is useful in analyzing small scale long-term behavior. | |||
===Page Rank=== | |||
Page Rank was the original ranking algorithm used by Google's search engine to rank web pages.<ref> | |||
http://ilpubs.stanford.edu:8090/422/ | |||
</ref> The algorithm was created by the founders of Google, Larry Page and Sergey Brin as part of Page's PhD thesis. When a query is entered in a search engine, there are a set of web pages which are matched by this query, but this set of pages must be ordered by their "importance" in order to identify the most meaningful results first. Page Rank is an algorithm which assigns importance to every web page based on the links in each page. | |||
==== Intuition ==== | |||
We can represent web pages by a set of nodes, where web links are represented as edges connecting these nodes. Based on our intuition, there are three main factors in deciding whether a web page is important or not. | |||
# A web page is important if many other pages point to it. | |||
# The more important a webpage is, the more weight is placed on its links. | |||
# The more links a webpage has, the less weight is placed on its links. | |||
====Modelling==== | |||
We can model the set of links as a N-by-N matrix L, where N is the number of web pages we are interested in: | |||
<math>L_{ij} = | |||
\left\{ | |||
\begin{array}{lr} | |||
1 : \text{if page j points to i}\\ | |||
0 : \text{otherwise} | |||
\end{array} | |||
\right. | |||
</math> | |||
The number of outgoing links from page j is | |||
<math>c_j = \sum_{i=1}^N L_{ij}</math> | |||
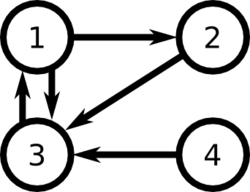
For example, consider the following set of links between web pages: | |||
[[File:PageRank.png|250px]] | |||
According to the factors relating to importance of links, we can consider two possible rankings : | |||
<math>\displaystyle 3 > 2 > 1 > 4 </math> | |||
or | |||
<math>\displaystyle 3>1>2>4 </math> | |||
if we consider that the high importance of the link from 3 to 1 is more influent than the fact that there are two outgoing links from page 1 and only one from page 2. | |||
We have <math>L = \begin{bmatrix} | |||
0 & 0 & 1 & 0 \\ | |||
1 & 0 & 0 & 0 \\ | |||
1 & 1 & 0 & 1 \\ | |||
0 & 0 & 0 & 0 | |||
\end{bmatrix}</math>, and <math>c = \begin{pmatrix}2 & 1 & 1 & 1\end{pmatrix} </math> | |||
We can represent the ranks of web pages as the vector P, where the i<sup>th</sup> element is the rank of page i: | |||
<math>P_i = (1-d) + d\sum_j \frac{L_{ij}}{c_j} P_j</math> | |||
Here we take the sum of the weights of the incoming links, where links are reduced in weight if the linking page has a lot of outgoing links, and links are increased in weight if the linking page has a lot of incoming links. | |||
We don't want to completely ignore pages with no incoming links, which is why we add the constant (1 - d). | |||
If | |||
<math>L = \begin{bmatrix} L_{11} & \cdots & L_{1N} \\ | |||
\vdots & \ddots & \vdots \\ | |||
L_{N1} & \cdots & L_{NN} \end{bmatrix}</math> | |||
<math>D = \begin{bmatrix} c_1 & \cdots & 0 \\ | |||
\vdots & \ddots & \vdots \\ | |||
0 & \cdots & c_N \end{bmatrix}</math> | |||
Then <math>D^{-1} = \begin{bmatrix} c_1^{-1} & \cdots & 0 \\ | |||
\vdots & \ddots & \vdots \\ | |||
0 & \cdots & c_N^{-1} \end{bmatrix}</math> | |||
<math>\!P = (1-d)e + dLD^{-1}P</math> | |||
where <math>\!e = \begin{pmatrix} 1 \\ \vdots \\ 1 \end{pmatrix}</math> is the vector with all 1's | |||
To simplify the problem, we let <math>\!e^T P = N \Rightarrow \frac{e^T P}{N} = 1</math>. This means that the average importance of all pages on the internet is 1. | |||
Then | |||
<math>\!P = (1-d)\frac{ee^TP}{N} + dLD^{-1}P</math> | |||
::<math>\! = \left [ (1-d)\frac{ee^T}{N} + dLD^{-1} \right ] P</math> | |||
::<math>\! = \left [ \left ( \frac{1-d}{N} \right ) E + dLD^{-1} \right ] P</math>, where <math> E </math> is an NxN matrix filled with ones. | |||
Let <math>\!A = \left [ \left ( \frac{1-d}{N} \right ) E + dLD^{-1} \right ]</math> | |||
Then <math>\!P = AP</math>. | |||
Note that P is a stationary distribution and, more importantly, P is an eigenvector of A with eigenvalue 1. Therefore, we can find the ranks of all web pages by solving this equation for P. | |||
We can find the vector P for the example above, using the following Matlab code: | |||
L = [0 0 1 0; 1 0 0 0; 1 1 0 1; 0 0 0 0]; | |||
D = [2 0 0 0; 0 1 0 0; 0 0 1 0; 0 0 0 1]; | |||
d = 0.8 ;% pages with no links get a weight of 0.2 | |||
N = 4 ; | |||
A = ((1-d)/N) * ones(N) + d * L * inv(D); | |||
[EigenVectors, EigenValues] = eigs(A) | |||
s=sum(EigenVectors(:,1));% we should note that the average entry of P should be 1 according to our assumption | |||
P=(EigenVectors(:,1))/s*N | |||
This outputs: | |||
EigenVectors = | |||
-0.6363 0.7071 0.7071 -0.0000 | |||
-0.3421 -0.3536 + 0.3536i -0.3536 - 0.3536i -0.7071 | |||
-0.6859 -0.3536 - 0.3536i -0.3536 + 0.3536i 0.0000 | |||
-0.0876 0.0000 + 0.0000i 0.0000 - 0.0000i 0.7071 | |||
EigenValues = | |||
1.0000 0 0 0 | |||
0 -0.4000 - 0.4000i 0 0 | |||
0 0 -0.4000 + 0.4000i 0 | |||
0 0 0 0.0000 | |||
P = | |||
1.4528 | |||
0.7811 | |||
1.5660 | |||
0.2000 | |||
Note that there is an eigenvector with eigenvalue 1. | |||
The reason why there always exist a 1-eigenvector is that A is a stochastic matrix. | |||
Thus our vector P is <math> | |||
\begin{bmatrix}1.4528 \\ 0.7811 \\ 1.5660\\ 0.2000 \end{bmatrix}</math> | |||
However, this method is not practical, because there are simply too many web pages on the internet. So instead Google uses a method to approximate an eigenvector with eigenvalue 1. | |||
Note that page three has the rank with highest magnitude and page four has the rank with lowest magnitude, as expected. | |||
==''' Markov Chain Monte Carlo - Metropolis-Hastings - October 25th, 2011'''== | |||
We want to find <math> \int h(x)f(x)\, \mathrm dx </math>, but we don't know how to sample from <math>\,f</math>. | |||
We have seen simple techniques earlier in the course; here is an example of a real life application. It consists of the search for a Markov Chain such that its stationary distribution is <math>\,f</math>. | |||
==== Main procedure ==== | |||
Let us suppose that <math>\,q(y|x)</math> is a friendly distribution: we can sample from this function. | |||
1. Initialize the chain with <math>\,x_{i}</math> and set <math>\,i=0</math>. | |||
2. Draw a point from <math>\,q(y|x)</math> i.e. <math>\,Y \backsim q(y|x_{i})</math>. | |||
3. Evaluate <math>\,r(x,y)=min\left\{\frac{f(y)}{f(x)}\frac{q(x|y)}{q(y|x)},1\right\}</math> | |||
4. Draw a point <math>\,U \backsim Unif[0,1]</math>. | |||
5. <math>\,x_{i+1}=\begin{cases}y & \text{ if } U<r \\x_{i} & \text{ otherwise } \end{cases} </math>. | |||
6. <math>\,i=i+1</math>. Go back to 2. | |||
==== Remark 1 ==== | |||
A very common choice for <math>\,q(y|x)</math> is <math>\,N(y;x,b^{2})</math>, a normal distribution centered at the current point. | |||
Note : In this case <math>\,q(y|x)</math> is symmetric i.e. <math>\,q(y|x)=q(x|y)</math>. | |||
(Because <math>\,q(y|x)=\frac{1}{\sqrt{2\pi}b}e^{-\frac{1}{2b^{2}}(y-x)^{2}}</math> and <math>\,(y-x)^{2}=(x-y)^{2}</math>). | |||
Thus we have <math>\,\frac{q(x|y)}{q(y|x)}=1</math>, which implies : | |||
<math>\,r(x,y)=min\left\{\frac{f(y)}{f(x)},1\right\}</math>. | |||
In general, if <math>\,q(x|y)</math> is symmetric then the algorithm is called Metropolis in reference to the original algorithm (made in 1953)<ref>http://en.wikipedia.org/wiki/Equations_of_State_Calculations_by_Fast_Computing_Machines</ref>. | |||
====Remark 2==== | |||
The value y is accepted if <math>\,u<min\left\{\frac{f(y)}{f(x)},1\right\}</math> so it is accepted with the probability <math>\,min\left\{\frac{f(y)}{f(x)},1\right\}</math>. | |||
Thus, if <math>\,f(y)>f(x)</math>, then <math>\,y</math> is always accepted. | |||
The higher that value of the pdf is in the vicinity of a point <math>\,y_1</math>, the more likely it is that a random variable will take on values around <math>\,y_1</math>. As a result it makes sense that we would want a high probability of acceptance for points generated near <math>\,y_1</math>. | |||
====Remark 3==== | |||
One strength of the Metropolis-Hastings algorithm is that normalizing constants, which are often quite difficult to determine, can be cancelled out in the ratio <math> r </math>. For example, consider the case where we want to sample from the beta distribution, which has the pdf: | |||
<math> | |||
\begin{align} | |||
f(x;\alpha,\beta)& = \frac{1}{\mathrm{B}(\alpha,\beta)}\, x^{\alpha-1}(1-x)^{\beta-1}\end{align} | |||
</math> | |||
The beta function, ''B'', appears as a normalizing constant but it can be simplified by construction of the method. | |||
====Example==== | |||
<math>\,f(x)=\frac{1}{\pi^{2}}\frac{1}{1+x^{2}}</math> | |||
Then, we have <math>\,f(x)\propto\frac{1}{1+x^{2}}</math>. | |||
And let us take <math>\,q(x|y)=\frac{1}{\sqrt{2\pi}b}e^{-\frac{1}{2b^{2}}(y-x)^{2}}</math>. | |||
Then <math>\,q(x|y)</math> is symmetric. | |||
Therefore Y can be simplified. | |||
We get : | |||
<math>\,\begin{align} | |||
\displaystyle r(x,y) | |||
& =min\left\{\frac{f(y)}{f(x)}\frac{q(x|y)}{q(y|x)},1\right\} \\ | |||
& =min\left\{\frac{f(y)}{f(x)},1\right\} \\ | |||
& =min\left\{ \frac{ \frac{1}{1+y^{2}} }{ \frac{1}{1+x^{2}} },1\right\}\\ | |||
& =min\left\{ \frac{1+x^{2}}{1+y^{2}},1\right\}\\ | |||
\end{align} | |||
</math>. | |||
The Matlab code of the algorithm is the following : | |||
<pre> | |||
clear all | |||
close all | |||
clc | |||
b=2; | |||
x(1)=randn; | |||
for i=2:10000 | |||
y=b*randn+x(i-1); | |||
r=min((1+x(i-1)^2)/(1+y^2),1); | |||
u=rand; | |||
if u<r | |||
x(i)=y; | |||
else | |||
x(i)=x(i-1); | |||
end | |||
end | |||
hist(x(5000:end)); | |||
%The Markov Chain usually takes some time to converge and this is known as the "burning time". | |||
%Therefore, we don't display the first 5000 points because they don't show the limiting behaviour of the Markov | |||
Chain. | |||
</pre> | |||
As we can see, the choice of the value of b is made by us. | |||
Changing this value has a significant impact on the results we obtain. There is a pitfall when b is too big or too small. | |||
Example with <math>\,b=0.1</math> (Also with graph after we run j=5000:10000; plot(j,x(5000:10000)): | |||
[[File:redaccoursb01.JPG|300px]] [[File:001Metr.PNG|300px]] | |||
With <math>\,b=0.1</math>, the chain takes small steps so the chain doesn't explore enough of the sample space. It doesn't give an accurate report of the function we want to sample. | |||
Example with <math>\,b=10</math> : | |||
[[File:redaccoursb10.JPG|300px]] [[File:010metro.PNG|300px]] | |||
With <math>\,b=10</math>, jumps are very unlikely to be accepted as they deviate far from the mean (i.e. <math>\,y</math> is rejected as <math>\ u>r </math> and <math>\,x(i)=x(i-1)</math> most of the time, hence most sample points stay fairly close to the origin. | |||
The third graph that resembles white noise (as in the case of <math>\,b=2</math>) indicates better sampling as more points are covered and accepted. For <math>\,b=0.1</math>, we have lots of jumps but most values are not repeated, hence the stationary distribution is less obvious; whereas in the <math>\,b=10</math> case, many points remains around 0. Approximately 73% were selected as x(i-1). | |||
Example with <math>\,b=2</math> : | |||
[[File:redaccoursb2.JPG|300px]] [[File:100metr.PNG|300px]] | |||
With <math>\,b=2</math>, we get a more accurate result as we avoid these extremes. Approximately 37% were selected as x(i-1). | |||
If the sample from the Markov Chain starts to look like the target distribution quickly, we say the chain is mixing well. | |||
==''' Theory and Applications of Metropolis-Hastings - October 27th, 2011'''== | |||
As mentioned in the previous section, the idea of the Metropolis-Hastings (MH) algorithm is to produce a Markov chain that converges to a stationary distribution <math>\!f</math> which we are interested in sampling from. | |||
====Convergence==== | |||
One important fact to check is that <math>\displaystyle f</math> is indeed a stationary distribution in the MH scheme. For this, we can appeal to the implications of the detailed balance property: | |||
Given a probability vector <math>\!\pi</math> and a transition matrix <math>\displaystyle P</math>, <math>\!\pi</math> has the detailed balance property if <math>\!\pi_iP_{ij} = P_{ji}\pi_j</math> | |||
If <math>\!\pi</math> satisfies detailed balance, then it is a stationary distribution. | |||
The above definition applies to the case where the states are discrete. In the continuous case, <math>\displaystyle f</math> satisfies detailed balance if <math>\displaystyle f(x)p(x,y)=f(y)p(y,x)</math>. Where <math>\displaystyle p(x,y)</math> and <math>\displaystyle p(y,x)</math> are the probabilities of transitioning from x to y and y to x respectively. If we can show that <math>\displaystyle f</math> has the detailed balance property, we can conclude that it is a stationary distribution. Because <math>\int^{}_y f(y)p(y,x)dy=\int^{}_y f(x)p(x,y)dy=f(x)</math>. | |||
In the MH algorithm, we use a proposal distribution to generate <math>\, y </math>~<math>\displaystyle q(y|x)</math>, and accept y with probability <math>min\left\{\frac{f(y)}{f(x)}\frac{q(x|y)}{q(y|x)},1\right\}</math> | |||
Suppose, without loss of generality, that <math>\frac{f(y)}{f(x)}\frac{q(x|y)}{q(y|x)} \le 1</math>. This implies that <math>\frac{f(x)}{f(y)}\frac{q(y|x)}{q(x|y)} \ge 1</math> | |||
Let <math>\,r(x,y)</math> be the chance of accepting point y given that we are at point x. | |||
So <math>\,r(x,y) = min\left\{\frac{f(y)}{f(x)}\frac{q(x|y)}{q(y|x)},1\right\} = \frac{f(y)}{f(x)} \frac{q(x|y)}{q(y|x)}</math> | |||
Let <math>\,r(y,x)</math> be the chance of accepting point x given that we are at point y. | |||
So <math>\,r(y,x) = min\left\{\frac{f(x)}{f(y)}\frac{q(y|x)}{q(x|y)},1\right\} = 1</math> | |||
<math>\,p(x,y)</math> is the probability of generating and accepting y, while at point x. | |||
So <math>\,p(x,y) = q(y|x)r(x,y) = q(y|x) \frac{f(y)}{f(x)} \frac{q(x|y)}{q(y|x)} = \frac{f(y)q(x|y)}{f(x)}</math> | |||
<math>\,p(y,x)</math> is the probability of generating and accepting x, while at point y. | |||
So <math>\,p(y,x) = q(x|y)r(y,x) = q(x|y)</math> | |||
<math>\,f(x)p(x,y) = f(x)\frac{f(y)q(x|y)}{f(x)} = f(y)q(x|y) = f(y)p(y,x)</math> | |||
Thus, detailed balance holds. | |||
:i.e. <math>\,f(x)</math> is stationary distribution | |||
It can be shown (although not here) that <math>\!f</math> is a limiting distribution as well. Therefore, the MH algorithm generates a sequence whose distribution converges to <math>\!f</math>, the target. | |||
====Implementation==== | |||
In the implementation of MH, the proposal distribution is commonly chosen to be symmetric, which simplifies the calculations and makes the algorithm more intuitively understandable. The MH algorithm can usually be regarded as a random walk along the distribution we want to sample from. Suppose we have a distribution <math>\!f</math>: | |||
[[File:Standard normal distribution.gif]] | |||
Suppose we start the walk at point <math>\!x</math>. The point <math>\!y_{1}</math> is in a denser region than <math>\!x</math>, therefore, the walk will always progress from <math>\!x</math> to <math>\!y_{1}</math>. On the other hand, <math>\!y_{2}</math> is in a less dense region, so it is not certain that the walk will progress from <math>\!x</math> to <math>\!y_{2}</math>. In terms of the MH algorithm: | |||
<math>r(x,y_{1})=min(\frac{f(y_{1})}{f(x)},1)=1</math> since <math>\!f(y_{1})>f(x)</math>. Thus, any generated value with a higher density will be accepted. | |||
<math>r(x,y_{2})=\frac{f(y_{2})}{f(x)}</math>. The lower the density of <math>\!y_{2}</math> is, the less chance it will have of being accepted. | |||
A certain class of proposal distributions can be written in the form: | |||
<math>\,y|x_i = x_i + \epsilon_i</math> | |||
where <math>\,\epsilon_i = g(|x-y|)</math> | |||
The density depends only on the distance between the current point and the next one (which can be seen as the "step" being taken). These proposal distributions give the Markov chain the random walk nature. The normal distribution that we frequently use in our examples satisfies the above definition. | |||
In actual implementations of the MH algorithm, the proposal distribution needs to be chosen judiciously, because not all proposals will work well with all target distributions we want to sample from. Take a trimodal distribution for example: | |||
[[File:trimodal.jpg]] | |||
If we choose the proposal distribution to be a standard normal as we have done before, problems will arise. The low densities between the peaks means that the MH algorithm will almost never walk to any points generated in these regions and get stuck at one peak. One way to address this issue is to increase the variance, so that the steps will be large enough to cross the gaps. Of course, in this case, it would probably be beneficial to come up with a different proposal function. As a rule of thumb, such functions should result in an approximately 50% acceptance rate for generated points. | |||
====Simulated Annealing==== | |||
Metropolis-Hastings is very useful in simulation methods for solving optimization problems. One such application is simulated annealing, which addresses the problems of minimizing a function <math>\!h(x)</math>. This method will not always produce the global solution, but it is intuitively simple and easy to implement. | |||
Consider <math>e^{\frac{-h(x)}{T}}</math>, maximizing this expression is equivalent to minimizing <math>\!h(x)</math>. Suppose <math>\mu</math> is the maximizing value and <math>h(x)=(x-\mu)^2</math>, then the maximization function is a gaussian distribution <math>\!e^{-\frac{(x-\mu)^2}{T}}</math>. When many samples are taken from this distribution, the mean will converge to the desired maximizing value. The annealing comes into play by lowering ''T'' (the temperature) as the sampling progresses, making the distribution narrower. The steps of simulated annealing are outlined below: | |||
1. start with a random <math>\!x</math> and set ''T'' to a large number | |||
2. generate <math>\!y</math> from a proposal distribution <math>\!q(y|x)</math>, which should be symmetric | |||
3. accept <math>\!y</math> with probability <math>min(\frac{f(y)}{f(x)},1)</math> . (Note: <math>f(x) = e^{\frac{-h(x)}{T}}</math>) | |||
4. decrease ''T'', and then go to step 2 | |||
To decrease ''T'' in Step 4, a variety of functions can be used. For example, a common temperature function used is with geometric decline, given by an initial temperature <math>\! T_o </math>, final temperature <math>\! T_f </math>, the number of steps n, and the temperature function <math>\ T(t) = T_0(\frac{T_f}{T_o})^{t/n} </math> | |||
The following plot and Matlab code illustrates the simulated annealing procedure as temperature ''T'', the variance, decreases for a Gaussian distribution with zero mean. Starting off with a large value for the temperature ''T'' allows the Metropolis-Hastings component of the procedure to capture the mean, before gradually decreasing the temperature ''T'' in order to converge to the mean. | |||
[[File:Simulated annealing illustration.png]] | |||
x=-10:0.1:10; | |||
mu=0; | |||
T=5; | |||
colour = ['b', 'g', 'm', 'r', 'k']; | |||
for i=1:5 | |||
pdfNormal=normpdf(x, mu, T); | |||
plot(x, pdfNormal, colour(i)); | |||
T=T-1; | |||
hold on | |||
end | |||
hleg1=legend('T=5', 'T=4', 'T=3', 'T=2', 'T=1'); | |||
title('Simulated Annealing Illustration'); | |||
=='''References'''== | |||
<references/> | |||
=='''Simulated Annealing and Gibbs Sampling - November 1, 2011'''== | |||
continued from previous lecture... | |||
Recall <math>\ r(x,y)=min(\frac{f(y)}{f(x)},1)</math> where <math> \frac{f(y)}{f(x)} = \frac{e^{\frac{-h(y)}{T}}}{e^{\frac{-h(x)}{T}}} = e^{\frac{h(x)-h(y)}{T}}</math> where <math>\ r(x,y) </math> represents the probability of accepting <math>\ y</math>. | |||
We will now look at a couple cases where <math> \displaystyle h(y) > h(x) </math> or <math> \displaystyle h(y) < h(x) </math>, and explore whether to accept or reject <math>\ y </math>. | |||
====Cases==== | |||
Case a) | |||
Suppose <math> \displaystyle h(y) < h(x) </math>. Since we want to find the minimum value for <math>\displaystyle h(x) </math>, and the point <math>\displaystyle y </math> creates a lower value than our previous point, we accept the new point. Mathematically, <math>\displaystyle h(y) < h(x) </math> implies that: | |||
<math> \frac{f(y)}{f(x)} > 1 </math>. Therefore, | |||
<math> \displaystyle r = 1 </math>. | |||
So, we will always accept <math>\displaystyle y </math>. | |||
Case b) | |||
Suppose <math> \displaystyle h(y) > h(x) </math>. This is bad, since our goal is to minimize <math>\displaystyle h(x) </math>. However, we may still accept <math>\displaystyle y </math> with some chance: | |||
<math> \frac{f(y)}{f(x)} < 1 </math>. Therefore, | |||
<math>\displaystyle r < 1 </math>. | |||
So, we may accept <math>\displaystyle y </math> with probability <math>\displaystyle r </math>. | |||
Next, we will look at these cases as <math>\displaystyle T\to0 </math>. | |||
As <math>\displaystyle T\to0 </math> and case a) happens, <math> e^{\frac{h(x)-h(y)}{T}} \to \infty </math>, so we will always accept <math>\displaystyle y </math>. | |||
As <math>\displaystyle T\to0 </math> and case b) happens, <math> e^{\frac{h(x)-h(y)}{T}} \to \ 0 </math>, so the probability that <math>\displaystyle y </math> will be accepted gets extremely small. | |||
It is worth noting that if we simply start with a small value of T, we may end up rejecting all the generated points, and hence we will get stuck somewhere in the function (due to case b)), which might be a maximum value in some intervals (local maxima), but not in the whole domain (global maxima). It is therefore necessary to start with a large value of T in order to explore the whole function. At the same time, a good estimation of <math>\ x_0 </math> < is needed (at least cannot differ from the maximum point too much). | |||
=====Example===== | |||
Let <math>\displaystyle h(x) = (x-2)^2 </math>. | |||
The graph of it is: | |||
[[File:PCh(x).jpg|center|500]] | |||
Then, <math> e^{\frac{-h(x)}{T}} = e^{\frac{-(x-2)^2}{T}} </math> . Take an initial value of T = 20. A graph of this is: | |||
[[File:PC-highT.jpg|center|500]] | |||
In comparison, we look a graph of T = 0.2: | |||
[[File:PC-lowT.jpg|center|500]] | |||
One can see that with a low T value, the graph has a lot of r = 0, and r >1, while having a bigger T value gives smoother transitions in the graph. | |||
The MATLAB code for the above graphs are: | |||
<pre> | |||
ezplot('(x-2)^2',[-6,10]) | |||
ezplot('exp((-(x-2)^2)/20)',[-6,10]) | |||
ezplot('exp((-(x-2)^2)/0.2)',[-6,10]) | |||
</pre> | |||
=====Travelling Salesman Problem===== | |||
The simulated annealing method can be applied to compute the solution to the travelling salesman problem. Suppose there are N cities and the salesman only have to visit each city once. The objective is to find out the shortest path (i.e. shortest total length of journey) connecting the cities. An algorithm using simulated annealing on the problem can be found here ([http://www.cs.ubbcluj.ro/~csatol/mestint/pdfs/Numerical_Recipes_Simulated_Annealing.pdf Reference]). | |||
===Gibbs Sampling=== | |||
Gibbs sampling is another Markov chain Monte Carlo method, similar to Metropolis-Hastings. There are two main differences between Metropolis-Hastings and Gibbs sampling. First, the candidate state is always accepted as the next state in Gibbs sampling. Second, it is assumed that the full conditional distributions are known, i.e. <math>P(X_i=x|X_j=x_j, \forall j\neq i)</math> for all <math>\displaystyle i</math>. The idea is that it is easier to sample from conditional distributions which are sets of one dimensional distributions than to sample from a joint distribution which is a higher dimensional distribution. Gibbs is a way to turn the joint distribution into multiple conditional distributions. | |||
<b>Advantages:</b><br /> | |||
- Sampling from conditional distributions may be easier than sampling from joint distributions | |||
<b>Disadvantages:</b><br /> | |||
- We do not necessarily know the conditional distributions | |||
For example, if we want to sample from <math>\, f_{X,Y}(x,y)</math>, we need to know how to sample from <math>\, f_{X|Y}(x|y)</math> and <math>\, f_{Y|X}(y|x)</math>. Suppose the chain starts with <math>\,(X_0,Y_0)</math> and <math>(X_1,Y_1), \dots , (X_n,Y_n)</math> have been sampled. Then, | |||
<math>\, (X_{n+1},Y_{n+1})=(f_{X|Y}(x|Y_n),f_{Y|X}(y|X_{n+1}))</math> | |||
Gibbs sampling turns a multi-dimensional distribution into a set of one-dimensional distributions. If we want to sample from | |||
<math>P_{X^1,\dots ,X^p}(x^1,\dots ,x^p)</math> | |||
and the full conditionals are known, then: | |||
<math>X^1_{n+1}=f(X^1|X^2_n,\dots ,X^p_n)</math> | |||
<math>X^2_{n+1}=f(X^2|X^1_{n+1},X^3_n\dots ,X^p_n)</math> | |||
<math>\vdots</math> | |||
<math>X^{p-1}_{n+1}=f(X^{p-1}|X^1_{n+1},\dots ,X^{p-2}_{n+1},X^p_n)</math> | |||
<math>X^p_{n+1}=f(X^p|X^1_{n+1},\dots ,X^{p-1}_{n+1})</math> | |||
With Gibbs sampling, we can simulate <math>\displaystyle n</math> random variables sequentially from <math>\displaystyle n</math> univariate conditionals rather than generating one <math>\, n </math>-dimensional vector using the full joint distribution, which could be a lot more complicated. | |||
Computational inference deals with probabilistic graphical models. Gibbs sampling is useful here: graphical models show the dependence relations among random variables. For instance, Bayesian networks are graphical models represented using directed acyclic graphs. Looking at such a graphical model tells us on which random variable the distribution of a certain random variable depends (i.e. its parent). The model can be used to "factor" a joint distribution into conditional distributions. | |||
[[File:stat341_nov_1_graphical_model.png|200px|thumb|left|Sample graphical model of five RVs]] | |||
For example, consider the five random variables A, B, C, D, and E. Without making any assumptions about dependence relations among them, all we know is | |||
<math>\, P(A,B,C,D,E)=</math><math>\, P(A|B,C,D,E) P(B|C,D,E) P(C|D,E) P(D|E) P(E)</math> | |||
However, if we know the relation between the random variables, e.g. given the graphical model on the left, we can simplify this expression: | |||
<math>\, P(A,B,C,D,E)=P(A) P(B|A) P(C|A) P(D|C) P(E|C)</math> | |||
Although the joint distribution may be very complicated, the conditional distributions may not be. | |||
Check out the following notes on Gibbs sampling: | |||
* [http://web.mit.edu/~wingated/www/introductions/mcmc-gibbs-intro.pdf MCMC and Gibbs Sampling, MIT Lecture Notes] | |||
* chapter 7.4 in [http://stat.fsu.edu/~anuj/pdf/classes/CompStatI09/BOOK.pdf Notes on Computational Methods in Statistics] | |||
* chapter 4.9 in [http://www.ma.hw.ac.uk/~foss/StochMod/Ross_S.pdf Introduction to Probability Models] by Sheldon Ross | |||
====Example of Gibbs sampling: Multi-variate normal==== | |||
We'd like to generate samples from a bivariate normal with parameters | |||
<math>\mu = \begin{bmatrix}1\\ 2 \end{bmatrix} = \begin{bmatrix}\mu_1 \\ \mu_2 \end{bmatrix}</math> | |||
and <math>\sigma = \begin{bmatrix}1 && 0.9 \\ 0.9 && 1 \end{bmatrix}= \begin{bmatrix}1 && \rho \\ \rho && 1 \end{bmatrix}</math> | |||
The conditional distributions of multi-variate normal random variables are also normal: | |||
<math>\, f(x_1|x_2)=N(\mu_1 + \rho(x_2-\mu_2), 1-\rho^2)</math> | |||
<math>\, f(x_2|x_1)=N(\mu_2 + \rho(x_1-\mu_1), 1-\rho^2)</math> | |||
In general, if the joint distribution has parameters | |||
<math>\mu = \begin{bmatrix}\mu_1 \\ \mu_2 \end{bmatrix}</math> and <math>\Sigma = \begin{bmatrix} \Sigma _{1,1} && \Sigma _{1,2} \\ \Sigma _{2,1} && \Sigma _{2,2} \end{bmatrix}</math> | |||
then the conditional distribution <math>\, f(x_1|x_2)</math> has mean <math>\, \mu_{1|2} = \mu_1 + \Sigma _{1,2}(\Sigma _{2,2})^{-1}(x_2-\mu_2)</math> and variance <math>\, \Sigma _{1|2} = \Sigma _{1,1}-\Sigma _{1,2}(\Sigma _{2,2})^{-1}\Sigma _{2,1}</math>. | |||
Thus, the algorithm for Gibbs sampling is: | |||
*1) Set i = 1 | |||
*2) Draw <math>X_1^{(i)}</math> ~ <math>N(\mu_1 + \rho(X_2^{(i-1)}-\mu_2), 1-\rho^2)</math> | |||
*3) Draw <math>X_2^{(i)}</math> ~ <math>N(\mu_2 + \rho(X_1^{(i)}-\mu_1), 1-\rho^2)</math> | |||
*4) Set <math>X^{(i)} = [X_1^{(i)}, X_2^{(i)}]^T</math> | |||
*5) Increment i, return to step 2. | |||
The Matlab code implementation of this algorithm is as follows:<br> | |||
mu = [1; 2]; | |||
sigma = [1 0.9; 0.9 1]; | |||
X(:,1) = [1; 2]; | |||
r = 1 - 0.9^2; | |||
for i = 2:2000 | |||
X(1,i) = 1 + 0.9*(X(2,i-1) - mu(2)) + r*randn; | |||
X(2,i) = 2 + 0.9*(X(1,i) - mu(1)) + r*randn; | |||
end | |||
plot(X(1,:),X(2,:),'.') | |||
Which gives the following plot: | |||
[[File:Gibbs_Example.jpg]] | |||
=='''Principal Component Analysis (PCA) - November 8, 2011'''== | |||
Principal Component Analysis is a 100 year old algorithm used for reducing the dimensionality of data. As the number of dimensions increase, the number of data points needed to sample accurately increase by an exponential factor. | |||
<math>\, x\in \mathbb{R}^D \rarr y\in \mathbb{R}^d</math> | |||
<math>\ d \le D </math> | |||
We want to transform <math>\, x</math> to <math>\, y</math> such that we reduce the dimensionality yet lose little information. Generally, variation in the data provides information. Thus we would like to reduce the dimensionality but keep as much variation, or information, as the original set of data. Also, covariance amongst the data reduces the amount of information we can infer from the data. Therefore, we would like to also reduce covariance when we reduce dimensionality. | |||
For example, consider dots in a three dimensional space. By unrolling the 2D manifold that they are on, we can reduce the data to 2D while losing little information. '''Note:''' This is not an application of PCA, but it simply illustrates one way we can reduce dimensionality. | |||
Principle Component Analysis allows us to reduce data to a linear subspace of its original space. It works best when data is in a lower dimensional subspace of its original space. | |||
==== Probabilistic View ==== | |||
We can see a data set <math>\, x</math> as a high dimensional random variable governed by a low dimensional random variable <math>\, y</math>. Given <math>\, x</math>, we are trying to estimate <math>\, y</math>. | |||
We can see this in 2D linear regression, as the locations of data points in a scatter plot are governed by its approximate linear regression. The subspace that we have reduced the data to here is in the direction of variation in the data. | |||
====Principal Component Analysis==== | |||
Principal component analysis is an orthogonal linear transformation on a data set. It associates the data coordinates with a new set of orthogonal vectors, each representing the direction of the maximum variance of the data. That is, the first principal component is the direction of the maximum variance, the second principal component is the direction of the maximum variance orthogonal to the first principal component, the third principal component is the direction of the maximum variance orthogonal to the first and second principal component and so on, until we have D principal components, where D is the dimension of the original data. | |||
Suppose we have data represented by <math>\, X \in \mathbb{R}^{D \times n} </math> | |||
Note that we are assuming that the data is mean centered. In other words, the average of every row is zero. If it isn't, we shift the data to have a mean of zero by subtracting the mean of every row from each <math>\ x </math> value in that row. This pre-processing step essentially alters the data such that each row in <math>\ X </math> includes only how the data differs from the mean of the sample, hence ensuring that the first PC describes the direction of maximum variance. | |||
To find the first principal component, we want to find a unit vector <math>\ W \in \mathbb{R}^{D} </math> that maximizes the variance of <math>\,W^TX</math>. We restrict <math>\,W</math> to unit vectors since we are only looking for the direction of the vector of maximum variation: the actual scale of it is unnecessary. So <math>\,W^TW = 1</math>. | |||
The variance of <math>\,W^TX</math> is <math>\,W^TSW</math> where <math>\,S</math> is the covariance matrix of X. | |||
<math>\, S = (X-\mu)(X-\mu)^T = XX^T </math>, since <math>\ \mu </math> is just the zero vector after we center the data around the mean. | |||
So we have to solve the problem | |||
<math>\, \text {Max } W^TSW \text{ such that } W^TW = 1</math> | |||
Using the method of Lagrange multipliers, we have | |||
<math>\,L(W, \lambda) = W^TSW - \lambda(W^TW - 1) </math> | |||
We set | |||
<math>\, \frac{\partial L}{\partial W} = 0 </math> | |||
Note that <math>\, W^TSW</math> is a quadratic form. So we have | |||
<math>\, \frac{\partial L}{\partial W} = 2SW - 2\lambda W = 0 </math> | |||
<math>\, SW = \lambda W </math> | |||
Since S is a matrix and lambda is a scaler, W is an eigenvector of S and lambda is its corresponding eigenvalue. | |||
Suppose that | |||
<math>\, \lambda_1 \ge \lambda_2 \ge \cdots \ge \lambda_D</math> | |||
are eigenvalues of S and <math>\, u_1, u_2, \cdots u_D</math> are their corresponding eigenvectors. | |||
We want to choose some <math>\, W = u </math> | |||
<math>\,u^TSu =u^T\lambda u = \lambda u^Tu = \lambda</math> | |||
So to maximize <math>\, u^TSu</math>, choose the eigenvector corresponding to the max eiegenvalue, e.g. <math>\, u_1</math>. | |||
So we let <math>\, W = u_1 </math> be the first principal component. | |||
The principal components decompose the total variance in the data. | |||
<math>\, \sum_{i=1}^D \text{Var}(u_i) = \sum_{i=1}^D \lambda_i = \text{Tr}(S) = \sum_{i=1}^D \text{Var}(x_i)</math> | |||
===Singular Value Decomposition=== | |||
Singular value decomposition is a "generalization" of eigenvalue decomposition "to rectangular matrices of size ''mxn''."<ref name="Abdel_SVD">Abdel-Rahman, E. (2011). Singular Value Decomposition [Lecture notes]. Retrieved from http://uwace.uwaterloo.ca</ref> Singular value decomposition solves:<br><br> | |||
:<math>\ A_{m\times n}\ v_{n\times 1}=s\ u_{m\times 1}</math><br><br> | |||
"for the right singular vector ''v'', the singular value ''s'', and the left singular vector ''u''. There are ''n'' singular values ''s''<sub>''i''</sub> and ''n'' right and left singular vectors that must satisfy the following conditions"<ref name="Abdel_SVD"/>: | |||
# "All singular values are non-negative"<ref name="Abdel_SVD"/>, <br> <math>\ s_i \ge 0.</math> | |||
# All "right singular vectors are pairwise orthonormal"<ref name="Abdel_SVD"/>, <br> <math>\ v_i^{T}v_j=\delta_{i,j}.</math> | |||
# All "left singular vectors are pairwise orthonormal"<ref name="Abdel_SVD"/>, <br> <math>\ u_i^{T}u_j=\delta_{i,j}.</math> | |||
where | |||
:<math>\delta_{i,j}=\left\{\begin{matrix}1 & \mathrm{if}\ i=j \\ 0 & \mathrm{if}\ i\neq j\end{matrix}\right.</math><br><br> | |||
'''Procedure to find the singular values and vectors'''<br> | |||
Observe the following about the eigenvalue decomposition of a real square matrix ''A'' where ''v'' is the unit eigenvector:<br> | |||
::<math> | |||
\begin{align} | |||
& Av=\lambda v \\ | |||
& (Av)^T=(\lambda v)^T \\ | |||
& (Av)^TAv=(\lambda v)^T\lambda v \\ | |||
& v^TA^TAv=\lambda^2v^Tv \\ | |||
& vv^TA^TAv=v\lambda^2 \\ | |||
& A^TAv=\lambda^2v | |||
\end{align} | |||
</math> | |||
As a result: | |||
# "The matrices ''A'' and ''A''<sup>''T''</sup>''A'' have the same eigenvectors."<ref name="Abdel_SVD"/> | |||
# "The eigenvalues of matrix ''A''<sup>''T''</sup>''A'' are the square of the eigenvalues of matrix ''A''."<ref name="Abdel_SVD"/> | |||
# Since matrix ''A''<sup>''T''</sup>''A'' is symmetric for any matrix A, | |||
## "all the eigenvalues of matrix ''A''<sup>''T''</sup>''A'' are real and distinct."<ref name="Abdel_SVD"/> | |||
## "the eigenvectors of matrix ''A''<sup>''T''</sup>''A'' are orthogonal and can be chosen to be orthonormal."<ref name="Abdel_SVD"/> | |||
# "The eigenvalues of matrix ''A''<sup>''T''</sup>''A'' are non-negative"<ref name="Abdel_SVD"/> since <math>\ \lambda^2_i \ge 0.</math> | |||
Conclusions 3 and 4 are "true even for a rectangular matrix ''A'' since ''A''<sup>''T''</sup>''A'' is still a square symmetric matrix"<ref name="Abdel_SVD"/> and its eigenvalues and eigenvectors can be found.<br><br> | |||
Therefore, for a rectangular matrix ''A'', assuming ''m>n'', the singular values and vectors can be found by: | |||
# "Form the ''nxn'' symmetric matrix ''A''<sup>''T''</sup>''A''."<ref name="Abdel_SVD"/> | |||
# Perform an eigenvalue decomposition to get ''n'' eigenvalues and their "corresponding eigenvectors, ordered such that"<ref name="Abdel_SVD"/> <br><math>\lambda_1 \ge \lambda_2 \ge \dots \ge \lambda_n \ge 0</math> and <math>\{v_1, v_2, \dots, v_n\}.</math> | |||
# "The singular values are"<ref name="Abdel_SVD"/>: <br><math>s_1=\sqrt{\lambda_1} \ge s_2=\sqrt{\lambda_2} \ge \dots \ge s_n=\sqrt{\lambda_n} \ge 0.</math><br>"The non-zero singular values are distinct; the equal sign applies only to the singular values that are equal to zero."<ref name="Abdel_SVD"/> | |||
# "The ''n''-dimensional right singular vectors are"<ref name="Abdel_SVD"/><br><math>\{v_1, v_2, \dots, v_n\}.</math> | |||
# "For the first <math>r \le n</math> singular values such that ''s''<sub>''i''</sub> ''> 0'', the left singular vectors are obtained as unit vectors"<ref name="Abdel_SVD"/> by <math>\tfrac{1}{s_i}Av_i=u_i.</math> | |||
# Select "the <math>\ m-r</math> left singular vectors corresponding to the zero singular values such that they are unit vectors orthogonal to each other and to the first ''r'' left singular vectors"<ref name="Abdel_SVD"/> <math>\{u_1, u_2, \dots, u_r\}.</math><br><br> | |||
'''Finding the Singular Value Decomposition Using MATLAB Code''' | |||
Please refer to the following link: http://www.mathworks.com/help/techdoc/ref/svd-singular-value-decomposition.html | |||
'''Formal definition'''<br> | |||
"We can now decompose the rectangular matrix ''A'' in terms of singular values and vectors as follows"<ref name="Abdel_SVD"/>:<br><br> | |||
<math>A_{m\times n} \begin{bmatrix} v_1 & | & \cdots & | & v_n \end{bmatrix}_{n\times n} = \begin{bmatrix} u_1 & | & \cdots & | & u_n & | I_{n+1} & | & \cdots & | & I_m \end{bmatrix}_{m\times m} \begin{bmatrix} s_1 & 0 & \cdots & 0 \\ 0 & s_2 & \cdots & 0 \\ \vdots & \vdots & \ddots & \vdots \\ 0 & 0 & \cdots & s_n \\ 0 & 0 & 0 & 0 \\ 0 & 0 & 0 & 0 \end{bmatrix}_{m\times n}</math> | |||
<br> | |||
Where <math>\ AV=US</math><br><br> | |||
Since "the matrices ''V'' and ''U'' are orthogonal"<ref name="Abdel_SVD"/>, ''V ''<sup>''-1''</sup>=''V''<sup>T</sup> and ''U ''<sup>''-1''</sup>=''U''<sup>T</sup>:<br><br> | |||
:<math>\ A=USV^T</math><br><br> | |||
"which is the formal definition of the singular value decomposition."<ref name="Abdel_SVD"/><br><br> | |||
'''Relevance to PCA'''<br> | |||
In order to perform PCA, one needs to do eigenvalue decomposition on the covariance matrix. By transforming the mean for all attributes to zero, the covariance matrix can be simplified to:<br><br> | |||
<math>\ S=XX^T</math><br><br> | |||
Since the eigenvalue decomposition of ''A''<sup>''T''</sup>''A'' gives the same eigenvectors as the singular value decomposition of ''A'', an additional and more consistent method (if a matrix has eigenvectors that are not invertible, its eigenvalue decomposition does not exist) for performing PCA is through the singular value decomposition of ''X''. | |||
The following MATLAB code uses singular value decomposition for performing PCA; 20 principal components, and thus the top 20 maximum variation directions, are selected for reconstructing facial images that have had noise applied to them: | |||
load noisy.mat | |||
%first noisy image; each image has a resolution of 20x28 | |||
imagesc(reshape(X(:,1),20,28)') | |||
%to grayscale | |||
colormap gray | |||
%singular value decomposition | |||
[u s v]=svd(X); | |||
%reduced feature space: 20 principal components | |||
Xh=u(:,1:20)*s(1:20,1:20)*v(:,1:20)'; | |||
figure | |||
imagesc(reshape(Xh(:,1),20,28)') | |||
colormap gray | |||
Since the reduced feature space image is noiseless, the added noise feature has less variation than the 20 principal components. | |||
=='''References'''== | |||
<references/> | |||
==''' PCA and Introduction to Kernel Function - November 10, 2011'''== | |||
(Continue from the last lecture) | |||
Some notations: | |||
Let <math>\displaystyle X</math> be a d by n matrix. | |||
Let <math>\displaystyle X_j\in\R^d</math> be the j th column of <math>\displaystyle X, \forall j=1,2,...,n</math>. | |||
Let <math>\displaystyle Q</math> be the covariance matrix of <math>\displaystyle X</math>. So <math>\displaystyle Q=(X_j-\bar{X})(X_j-\bar{X})^T</math>, where <math> \bar{X}=\frac{1}{n}\sum_{j=1}^n X_j</math>. Strictly speaking, <math>\displaystyle Q</math> is only the observed covariance matrix from the data collected as the real covariance matrix is unknown. | |||
But we are assuming that we have already centered the data, which means that <math>\bar{X}=0</math>. | |||
So <math>\displaystyle Q_{ij}=(X_i)(X_j)^T=[X X^T]_{ij} </math>. | |||
Now the principal components of <math>\displaystyle X</math> are the eigenvectors of <math>\displaystyle Q</math>. If we do the singular value decomposition, setting <math>\ [u\ s\ v] = svd(Q) </math>, then the columns of <math>\ u </math> are eigenvectors of <math>\displaystyle Q=X X^T</math>. | |||
Let <math>\displaystyle u</math> be a <math>\displaystyle d\times p</math> matrix, where <math>\displaystyle p < d</math>, that is composed of the <math>\ p </math> eigenvectors of <math>\displaystyle Q</math> corresponding to the largest eigenvalues. | |||
To map the data in a lower dimensional space we project <math>\displaystyle X</math> onto the <math>\ p </math> dimensional subspace defined by the columns of <math>\displaystyle u</math>, which are the first <math>\ p </math> principal components of <math>\displaystyle X</math>. | |||
Let <math>\displaystyle Y_{p\times n}={u^T}_{p\times d} X_{d\times n}</math>. So <math>\displaystyle Y_{p\times n}</math> is a lower dimensional approximation of our original data <math>\displaystyle X_{d\times n}</math> | |||
We can also approximately reconstruct the original data using the dimension-reduced data. However, we will lose some information because when we map those points into lower dimension, we throw away the last <math>\ (d - p) </math> eigenvectors which contain some of the original information. | |||
<math>\displaystyle X'_{d\times n} = {u}_{d\times p} Y_{p\times n}</math>, where <math>\displaystyle X'_{d\times n}</math> is an approximate reconstruction of <math>\displaystyle X_{d\times n}</math>. | |||
===Example Using Handwritten 2s and 3s=== | |||
The data X is a 64 by 400 matrix. Every column represents an 8 by 8 image of either a handwritten "2" or "3". The first 200 columns are 2s and the last 200 columns are 3s. First we center the data, and then we find the first 2 eigenvectors of <math>\displaystyle XX^T</math> using singular value decomposition. Finally we calculate <math>\displaystyle Y = u^TX</math> and plot the 200 data points in <math>\displaystyle Y</math> | |||
'''MATLAB CODE:''' | |||
MU = repmat(mean(X,2),1,400); | |||
% mean(X,2) is the average of all the rows stored in a column vector. | |||
% In order to center the data, we extend mean(X,2), which is a 64 by 1 matrix, into a 64 by 400 matrix. | |||
Xt = X-MU; | |||
% We have modified the data to zero mean data by subtracting the average of each row from every entry in that row. | |||
[u s v] = svd(Xt); | |||
% Note that size(u) == 64*64, and the columns of u are eigenvectors of VCM. | |||
Y = u(:,1:2)'*X; | |||
% We project X onto the subspace defined by the first two PCs to get Y. | |||
% This transforms the high dimensional points to lower 2 dimensional ones. | |||
plot(Y(1,1:200)',Y(2,1:200)','d') | |||
hold on | |||
plot(Y(1,201:400)',Y(2,201:400)','ro') | |||
% We now plot the lower dimensional projection of X. | |||
% Essentially we are plotting each point based on the magnitude of Principle Component #1 | |||
% and Principle Component #2 in that point. | |||
% Note that the first 200 columns represent 2s and are recorded by blue diamonds. | |||
% Note that the next 200 columns represent 3s and are recorded with red "o"s. | |||
The result is as follows, we can see clearly there are two classes - the 2s and 3s are generally divided into two sections: | |||
[[file:pca3.png|350px|400px]] | |||
In order to analyze the projection in more detail we can plot the original images on the graph. | |||
image = reshape(X,8,8,400); | |||
plotdigits(image,Y,.1,1); | |||
The result can now be seen more clearly from the following picture: | |||
[[file:Pca.png|350px|400px]] | |||
By examining this plot we can infer the approximate "meaning" of the first two principal components in this data. For instance, the points at the top of the plot tend to be slanted to the right, while the ones at the bottom are slanted to the left. So the second principal component quantifies the amount of slant in the number. | |||
===Introduction to Kernel Function=== | |||
PCA is useful when those data points spread in or close to a plane. This means that PCA is powerful when dealing with linear problems. But when data points spread in a manifold space, PCA is hard to implement. But there is a solution to this problem - we can use a method to change the linear algorithm into a nonlinear one. This is called the "Kernel Trick". | |||
'''An intuitive example''' | |||
[[File:Kernel trick.png|400px|300px]] | |||
From the picture, we can see the red circles are in the middle of the blue Xs. However, it is hard to separate those two classes by using any linear function (lines in the two dimensional space). But we can use a Kernal function <math>\ \phi</math> to project the points onto a three dimensional space. Once the blue Xs and red circles have been mapped to a three dimensional space in this way it is easy to separate them using a linear function. | |||
For more details about this trick, please see http://omega.albany.edu:8008/machine-learning-dir/notes-dir/ker1/ker1.pdf | |||
The significance of a Kernel Function, <math>\displaystyle \phi</math>, is that we can implicitly change the data into a high dimension. | |||
Let's look at how this is possible: | |||
<math>Z_1= | |||
\begin{bmatrix} | |||
x_1\\ | |||
y_1 | |||
\end{bmatrix}\xrightarrow{\phi} | |||
</math> | |||
<math>\phi(Z_1)= | |||
\begin{bmatrix} | |||
x_1^2\\ | |||
y_1^2\\ | |||
\sqrt2x_1y_1 | |||
\end{bmatrix} | |||
</math> | |||
<math>Z_2= | |||
\begin{bmatrix} | |||
x_2\\ | |||
y_2 | |||
\end{bmatrix}\xrightarrow{\phi} | |||
</math> | |||
<math>\phi(Z_2)= | |||
\begin{bmatrix} | |||
x_2^2\\ | |||
y_2^2\\ | |||
\sqrt2x_2y_2 | |||
\end{bmatrix} | |||
</math> | |||
The inner product of <math>\displaystyle \phi(Z1)</math> and <math>\displaystyle\phi(Z2)</math>, which is denoted as <math>\displaystyle\phi(Z1)^T\phi(Z2)</math>, is equal to: | |||
<math> | |||
\begin{bmatrix} | |||
x_1^2&y_1^2&\sqrt2x_1y_1 | |||
\end{bmatrix} | |||
\begin{bmatrix} | |||
x_2^2\\ | |||
y_2^2\\ | |||
\sqrt2x_2y_2 | |||
\end{bmatrix}=</math> <math>\displaystyle (x_1x_2+y_1y_2)^2=(Z_1^TZ_2)^2=<Z_1,Z_2>^2=K(Z_1,Z_2)</math>. | |||
'''The most common Kernel functions are as follows:''' | |||
*Linear: <math>\displaystyle K_{ij}=<X_i,X_j></math> | |||
*Polynomial:<math>\displaystyle K_{ij}=(1+<X_i,X_j>)^p</math> | |||
*Gaussian:<math>\displaystyle K_{ij}=e^\frac{-{\left\Vert X_i-X_j\right\|}^2}{2\sigma^2}</math> | |||
*Exponential:<math>\displaystyle K_{ij}=e^\frac{-{\left\Vert X_i-X_j\right\|}}{2\sigma^2}</math> | |||
Note: <math>{\left\Vert X_i-X_j\right\|}</math> denotes the distance between <math>\displaystyle X_i</math> and <math>\displaystyle X_j</math>. | |||
===Disadvantages of the Kernel Trick=== | |||
Since the kernel trick brings a lower dimensional problem to a higher dimensional space, it is affected by the curse of dimensionality. The curse of dimensionality states that it takes exponentially more points to estimate a solution in higher dimensions. ex) if <math>\R^1</math> needs 10 points, then <math>\R^2</math> needs 100, <math>\R^3</math> needs 1000 etc. | |||
==''' Kernel PCA - November 15, 2011'''== | |||
PCA doesn't work well when the directions of variation in our data are nonlinear. Especially in the case where the dataset has very high dimensions, the dataset lies near or on a nonlinear manifold which prevents PCA from determining principal components correctly. To deal with this problem, we apply kernels to PCA. By transforming the original data with a nonlinear mapping, we can obtain much better principal components. | |||
First we look at the algorithm for PCA and see how we can kernelize PCA: | |||
== PCA == | |||
Find eigenvectors of <math> \ XX^T </math>, call it <math>\ U </math> | |||
to map data points to a lower dimensional space:<br> | |||
<math>\ Y = U^{T}X</math><br> | |||
to reconstruct points:<br> | |||
<math>\ \hat{X} = UY</math><br> | |||
to map a new point:<br> | |||
<math>\ y = U^{T}x</math><br> | |||
to reconstruct point:<br> | |||
<math>\ \hat{x} = Uy</math><br> | |||
== Dual PCA == | |||
Consider the singular value decomposition of n-by-m matrix X: | |||
<math> | |||
\begin{align} | |||
\left[ U\ \Sigma\ V \right] & = svd(X) \\ | |||
X & = U\Sigma{V^T} | |||
\end{align} | |||
</math> | |||
Where: | |||
* The columns of U are the eigenvectors of <math>XX^T</math> corresponding to the eigenvalues in decreasing order. | |||
* The columns of V are the eigenvectors of <math>X^T{X}</math>. | |||
* The diagonal matrix <math>\Sigma</math> contains the square roots of the eigenvalues of <math>XX^T</math> in decreasing order. | |||
Now we want to kernelize this classical version of PCA. We would like to express everything based on V which can be kernelized. The reason why we want to do this is because if n >> m (i.e. the feature space is much larger than the number of sample points) then the original PCA algorithm would be impractical. We want an algorithm that depends less on n. This is called Dual PCA. | |||
U in terms of V: | |||
<math> | |||
\begin{align} | |||
X &= U\Sigma V^T \\ | |||
XV &= U\Sigma V^T V = U\Sigma \\ | |||
XV\Sigma^{-1} &= U\Sigma\Sigma^{-1} = U \\ | |||
U &= XV\Sigma^{-1} | |||
\end{align} | |||
</math> | |||
Y in terms of V: | |||
<math> | |||
\begin{align} | |||
X&=U \Sigma V^T \\ | |||
U^TX &= U^TU\Sigma V^T \\ | |||
U^TX &= \Sigma V^T \\ | |||
Y&=\Sigma V^T \\ | |||
\end{align} | |||
</math> | |||
Reconstructed points in terms of V: | |||
<math> | |||
\begin{align} | |||
\hat{X}&=UY \\ | |||
&=XV\Sigma^{-1}\Sigma{V^T} \\ | |||
&= XVV^T \\ | |||
&= X | |||
\end{align} | |||
</math> | |||
The value of y, a single point in the sample reduced to low-dimensional space, in terms of V: | |||
<math> | |||
\begin{align} | |||
y &= U^Tx \\ | |||
& = (XV\Sigma^{-1})^Tx \\ | |||
& = (\Sigma^{-1})^T{V^T}{x^T}x \\ | |||
& = \Sigma^{-1}{V^T}{x^T}x | |||
\end{align} | |||
</math> | |||
A single reconstructed point from the sample in terms of V: | |||
<math> | |||
\begin{align} | |||
\hat{x} &= Uy \\ | |||
& = UU^Tx \\ | |||
& =XV\Sigma^{-1}\Sigma^{-1}V^T{x^T}x \\ | |||
&= XV\Sigma^{-2}V^T{x^T}x | |||
\end{align} | |||
</math> | |||
===Kernel PCA=== | |||
The nonlinear mapping <math>\displaystyle\phi</math> allows for very high dimensional spaces and is never calculated explicitly. | |||
:<math> k(\mathbf{x},\mathbf{x}) = \phi^T(\mathbf{x})\phi(\mathbf{x})</math> | |||
<math> X_{d\times n} </math>, | |||
<math> X^TX_{n\times n} </math>, and | |||
<math> K(X,X)_{n\times n}</math> | |||
could have many different kernels. | |||
Generally, we want to replace <math> X^TX </math> with a kernel. The idea in Kernel PCA is that instead of finding the eigenvectors of <math> X^TX </math>, we can find the eigenvectors of a kernel. | |||
Example.<math>\displaystyle K(x_1,x_2)=e^\frac{-(X_1-X_2)^2}{\sigma}</math> | |||
Find the eigenvectors of the matrix | |||
<math>K=\left(\begin{matrix}0&0\dots&\dots\\ | |||
\vdots&\ddots\\\vdots\\\end{matrix}\right)</math> | |||
However, since the <math>\displaystyle\phi</math> is never calculated explicitly, kernel PCA cannot reconstruct points using the equation <math>\hat{X} = XVV^T</math> where <math> X=\phi</math>. | |||
In kernel PCA, we replace <math> k </math> for <math> \phi(X)^T \phi(X)</math>. This is correct if <math> \phi(X)</math> has a mean of zero. | |||
We need to find a way to centralize <math>\phi(X)</math>. | |||
===Centralizing Kernel PCA=== | |||
Recall in regular PCA, our variance-covariance matrix <math>\Sigma</math> was defined as <math>\Sigma = (X-\mu)(X-\mu)^T</math>. | |||
With kernel PCA, we will never explicitly define <math>\phi(x)</math> so we need a method to calculate its mean without calculating the actual transformation of <math>\phi(x)</math> itself. | |||
We define: | |||
:<math>\tilde{\phi}(x) = \phi(x) - E_x[\phi(x)]</math> | |||
And thus, our kernel function becomes: | |||
:<math>\tilde{k}(\mathbf{x},\mathbf{y}) = \tilde{\phi}(\mathbf{x})^T\tilde{\phi}(\mathbf{y})</math> | |||
:::<math>\! = (\phi(x) - E_x[\phi(x)])^T(\phi(y) - E_y[\phi(y)])</math> | |||
:::<math>\! = \phi(x)^T\phi(y) - \phi(y)E_x[\phi(x)^T] - \phi(x)^TE_y[\phi(y)] + E_x[\phi(x)^T]E_y[\phi(y)]</math> | |||
:::<math>\! = k(\mathbf{x},\mathbf{y}) - E_x[k(\mathbf{x},\mathbf{y})] - E_y[k(\mathbf{x},\mathbf{y})] + E_x[E_y[k(\mathbf{x},\mathbf{y})]]</math> | |||
In practice, we would do the following: | |||
1. Start with our matrix of data: <math> X_{d\times n} </math> | |||
2. Choose a kernel function <math> k </math> | |||
3. Compute <math>K</math>, an nxn matrix: <math>K=\left(\begin{matrix}k(x_1,x_1)&k(x_1,x_2)\dots&k(x_1,x_n)\\ | |||
\vdots&\ddots&\vdots\\k(x_n,x_1)&\dots&k(x_n,x_n)\\\end{matrix}\right)</math> | |||
4. Find <math>\tilde{K} = K - \frac{1}{n}\sum_{i=1}^{n}K(:,i) - \frac{1}{n}\sum_{j=1}^{n}K(j,:) + \frac{1}{n^2}\sum_{i=1}^{n}\sum_{j=1}^{n}K(i,j)</math> | |||
'''Note:''' On the right hand side, the second item is the average of the columns of <math>K</math>, the third item is the average of the rows of <math>K</math>, and the last item is the average of all the entries of <math>K</math>. | |||
As a result, they are vectors of dimension <math>1\times n</math>, <math>n\times 1</math>, and <math>1\times 1</math> (a scalar) respectively. To do the actual arithmetic, all matrix dimensions must match. In MATLAB, this is accomplished through the repmat() function. | |||
5. Find the eigenvectors of <math>\tilde{K}</math>. Take the first few and combine them in a matrix <math>V</math>, and use that as the <math>V</math> in the dual PCA mapping and reconstruction steps shown above. | |||
=='''Multidimensional Scaling (MDS)'''== | |||
Most of the common linear and polynomial kernels do not give very good results in practice. For real data, there is an alternate approach that can yield much better results but, as we will see later, is equivalent to a kernel PCA process. | |||
Multidimensional Scaling (MDS) is an algorithm, much like PCA, that maps the original high dimensional space to a lower dimensional space. | |||
===Introduction to MDS=== | |||
The main purpose of MDS is to try to map high dimensional data to a lower dimensional space while preserving the pairwise distances between points. | |||
ie: MDS addresses the problem of constructing a configuration of <math> \ n </math> points in Euclidean space by using information about the distance between the <math> \ n </math> patterns. | |||
It may not be possible to preserve the exact distances between points, so in this case we need to find a representation that is as close as possible. | |||
'''Definition''': A <math> \ n\times n </math> matrix <math> \ D</math> is called the '''distance matrix''' if: | |||
<math> \ D</math> is symmetric and its entries <math> \ d_{ij}</math> have the properties: | |||
<math> \ d_{ii} = 0</math> and <math> \ d_{ij} > 0</math> <math>\forall i\neq j </math> | |||
Given a distance matrix <math> \ D^{(X)}</math>, MDS attempts to find <math> \ n </math> data points <math> \ y_1,...,y_n</math> in <math> \ d </math> dimensions, such that if <math> \ d_{ij}^{(Y)}</math> denotes the Euclidean distance between <math> \ y_i, y_j</math> then <math> \ D^{(Y)}</math> is similar to <math> \ D^{(X)}</math>. | |||
===Metric MDS=== | |||
One of the possible methods to preserve the pairwise distance is to use Metric MDS, which attempts to minimize: | |||
<math>\min_Y \sum_{i=1}^{n}\sum_{j=1}^{n}(d_{ij}^{(X)} - d_{ij}^{(Y)})^2 </math> | |||
where <math>d_{ij}^{(X)} = ||x_i-x_j||^2 </math> and <math>d_{ij}^{(Y)} = ||y_i-y_j||^2 </math> | |||
==''' Continued with MDS, Isomap and Classification - November 22, 2011'''== | |||
<br />The distance matrix can be converted to a kernel matrix of inner products <math>\ X^TX</math> by <math>\ X^{T}X=-\frac{1}{2}HD^{(X)}H </math>,<br /> | |||
where <math>H=I-\frac{1}{n}ee^{T}</math> and <math>\ e </math> is a column vector of all 1. | |||
<br /><math> | |||
e=\left[\begin{array}{c} | |||
1\\ | |||
\vdots\\ | |||
1 | |||
\end{array}\right]_{(n\times1)} | |||
</math> | |||
<math>ee^{T}=\left[\begin{array}{ccc} | |||
1 & \cdots & 1\\ | |||
\vdots & \ddots & \vdots\\ | |||
1 & \cdots & 1 | |||
\end{array}\right]_{(n\times n)} | |||
</math> | |||
<math> | |||
H=\left[\begin{array}{ccc} | |||
1 & \cdots & 0\\ | |||
\vdots & \ddots & \vdots\\ | |||
0 & \cdots & 1 | |||
\end{array}\right]-\frac{1}{n}\left[\begin{array}{ccc} | |||
1 & \cdots & 1\\ | |||
\vdots & \ddots & \vdots\\ | |||
1 & \cdots & 1 | |||
\end{array}\right] | |||
</math> | |||
Now <math>\min_Y \sum_{i=1}^{n}\sum_{j=1}^{n}(d_{ij}^{(X)} - d_{ij}^{(Y)})^2 </math> can be reduced to <math>\min_Y \sum_{i=1}^{n}\sum_{j=1}^{n}(x_{i}^{T}x_{j}-y_{i}^{T}y_{j})^{2}</math> <math>\Leftrightarrow </math><math>\min_Y \ Tr(X^{T}X-Y^{T}Y)^{2}</math>. | |||
So far we have rewritten the objective function, but we must remember that our definition of <math>\ D </math> introduces constraints on its components. We must ensure that these constraints are respected in the way we have expressed the minimization problem with traces. Luckily, we can capture these constraints with a single requirement thanks to the following theorem: | |||
<br />''' Theorem''': Let <math>\ D</math> be a distance matrix and <math>\ K=X^{T}X=-\frac{1}{2}HD^{(X)}H </math>. Then <math>\ D</math> is Euclidean if and only if <math>\ K</math> is a positive semi-definite matrix. | |||
Therefore, to complete the rewriting of our original minimization problem from norms to traces, it suffices to impose that <math>\ K</math> be <math>\ p.s.d.</math>, as this guarantees that <math>\ D</math> is a distance matrix and the components of <math>\ X^{T}X </math> and <math>\ Y^{T}Y</math> satisfy the original constraints. | |||
<br />Proceeding with singular value decomposition, <math>\ X^{T}X</math> and <math>\ Y^{T}Y</math> can be decomposed as: | |||
<br /><math>\ X^{T}X=V\Lambda V^{T}</math><br /><math>Y^{T}Y=Q\hat{\Lambda} Q^{T}</math> | |||
<br />Since <math>\ Y^{T}Y</math> is <math>\ p.s.d.</math> , <math>\hat{\Lambda} </math> has no negative value and therefore: <math> Y=\hat{\Lambda}^{\frac{1}{2}}Q^{T}</math>. | |||
<br />The above definitions help rewrite the cost function as: | |||
<br /><math>\min_{Q,\hat{\Lambda}}Tr(V\Lambda V^{T}-Q\hat{\Lambda}Q^{T})^{2}</math> | |||
<br />Multiply <math>\ V^{T}</math> on the left and <math>\ V</math> on the right, we will get | |||
<br /><math>\min_{Q,\hat{\Lambda}}Tr(\Lambda-V^{T}Q\hat{\Lambda} Q^{T}V)^{2}</math> | |||
Then let <math>\ G=V^{T}Q</math> | |||
We can rewrite the target function as: | |||
<br /><math>\min_{Q,\hat{\Lambda}}Tr(\Lambda-G\hat{\Lambda}G^{T})^{2}</math> | |||
<math>=</math><math>\min_{G,\hat{\Lambda}}Tr(\Lambda^{2}+G{\Lambda}G^{T}G\hat{\Lambda}G^{T}-2\Lambda G\hat{\Lambda}G)</math> | |||
<br />For a fixed <math>\hat{\Lambda}</math> we can minimize for G. The result is that <math>\ G=I</math>. Then we can simplify the target function: | |||
<math>\min_{\hat{\Lambda}}Tr(\Lambda^{2}+\hat{\Lambda}^{2}-2\Lambda\hat{\Lambda}) | |||
</math> <math>=</math> <math>\min_{\hat{\Lambda}}Tr(\Lambda-\hat{\Lambda})^{2}</math> | |||
Obviously, <math>\hat{\Lambda}=\Lambda</math>. | |||
If <math>\ X</math> is <math>d\times n</math> matrix, <math>\ (d<n)</math>, the rank of <math>\ X</math> is no greater than <math>\ d</math>. Then the rank of <math>X^{T}X_{(n\times n)}</math> is no greater than <math>\ d</math> since <math>\ rank(X^{T}X) = rank(X) </math>. The dimension of <math>\ Y</math> is smaller than <math>\ X</math>, so therefore the rank of <math>\ Y^{T}Y</math> is smaller than <math>\ d</math>. | |||
Since we want to do dimensionality reduction and make <math>\ \Lambda</math> and <math>\hat{\Lambda}</math> as similar as possible, we can let <math>\hat{\Lambda}</math> be the top <math>\ d</math> diagonal elements of <math>\ \Lambda</math>. | |||
We also have <math>\ G = V^{T}Q </math>, obviously <math>\ Q=V</math> since <math>\ G = I</math>. | |||
The solution is: | |||
<br /><math>Y=\Lambda^{\frac{1}{2}}V^{T}</math> | |||
<br />where <math>\ V</math> is the eigenvector of <math>\ X^{T}X</math> corresponding to the top <math>\ d</math> | |||
eigenvalues, and <math>\Lambda</math> is the top <math>\ d</math> eigenvalues of <math>\ X^{T}X</math>. | |||
<br />Compare this with dual PCA. | |||
<br />In dual PCA, the result is | |||
<br /><math>\ Y=\Sigma V^{T}</math>. | |||
<br />Clearly, the result of dual PCA is the same with MDS. Actually, one property of PCA is to preserve the pairwise distances between data points in both high dimensional and low dimensional space. | |||
Now as an appendix, we provide a short proof to the first equation <math>\ X^{T}X=-\frac{1}{2}HD^{(X)}H </math>. | |||
<br />Let <math>\ S </math> be a vector where <math> S_{i}=X_i^TX_i,D=Se^T+eS^T-2X^TX </math>. | |||
<br />First notice <math>\ Xe=0 </math> as data are centered. | |||
<br /><math> HD^{(X)}H=(I-\frac{1}{n}ee^{T})(Se^T+eS^T-2X^TX)(I-\frac{1}{n}ee^{T}) </math> | |||
<br /><math> =-2X^TX+\frac{1}{n}ee^{T}2X^TX+\frac{1}{n}2X^TXee^{T}-\frac{1}{n^2}ee^{T}2X^TXee^{T} </math> | |||
<br /><math>\ =-2X^TX </math>, | |||
<br />as all other terms contains <math>\ Xe </math> or <math>\ e^TX^T </math>. | |||
<br />Hence, <math>\ X^{T}X=-\frac{1}{2}HD^{(X)}H </math>. | |||
===Isomap (As per Handout - Section 1.6)=== | |||
The Isomap algorithm is a nonlinear generalization of classical MDS with the main idea being that MDS is perfomed on the geodesic space of the non-linear data manifold as opposed to being performed on the input space. Isomap applies geodesic distances in the distance matrix for MDS rather than the straight line distances between the 2 points in order to find the low-dimensional mapping that preserves the pairwise distances. This geodesic distance is approximated by building a k-neighbourhood graph of all the points on the manifold and finding the shortest path to a given point. This gives a much better distance measurement in a manifold such as the 'Swiss Roll' manifold than using Euclidean distances. | |||
<br> | |||
'''Note''': Geodesic distance is the shortest path along the curved surface of the manifold measured as if the surface was flat | |||
Similarly to LLE, the Isomap algorithm proceeds in three steps: | |||
1. Finding the neighbours of each data points in high-dimensional data space | |||
2. Compute the geodesic pairwise distance between all points | |||
3. Integrate the data via MDS in order to preserve the distances | |||
<br>'''Process:'''<br> | |||
1. Identify the k nearest neighbours or choose points from a fixed radius | |||
2. Neighbour relations are represented by a graph G conneted with edges of weights <math> \ d_{X}(i,j)</math> | |||
3. The geodesic distances, <math> \ d_{M}(i,j)</math> between all pair points on the manifold, M, are then estimated | |||
'''Note''': Isomap approximates <math> \ d_{M}(i,j)</math> as the shortest path distance <math> \ d_{G}(i,j)</math> in Graph G. The k in the algorithm must be chosen carefully, too small a k and the graph will not be connected, but to large a k, and the algorithm will be closer to the euclidean distance instead. | |||
<br> | |||
4. Isomap applies classical MDS to <math> \ D^{(G)}</math> to generate an embedding of the data in d-dimensional Euclidean space Y | |||
<br> | |||
===Classification === | |||
Classification is a technique in pattern recognition. It can yield very complex decision boundaries as they are very suitable for [http://en.wikipedia.org/wiki/Data_synchronization#Ordered_data, ordered data], [http://en.wikipedia.org/wiki/Categorical_data, categorical data] or a mixture of the two types. A decision or classification represents a multi-stage decision process where a binary decision is made at each stage. | |||
'''E.g''': A hand-written object can be scanned and recognized by the '''classification''' technique. The model realized the class it belongs to and pairs it with the corresponding object in its library.<br> | |||
Mathematically, each object have a set of features <math>\ X</math> and a corresponding label <math>\ Y</math> which is the class it belongs to:<br> | |||
<math>\ \{(x_1,y_1),(x_2,y_2),...,(x_n,y_n)\}</math> | |||
Since the training set is labelled with the correct answers, classification is called a "'''supervised learning'''" method. | |||
In contrast, the '''Clustering''' technique is used to explore a data set whereby the main objective is to separate the sample into groups or to provide an understanding about the underlying structure or nature of the data. Clustering is an "'''unsupervised classification'''" method, because we do not know the groups that are in the data or any group characteristics of any unit observation. There are no labels to classify the data points, all we have is the feature set <math>\ X</math>:<br> | |||
<math>\ \{(x_1),(x_2),...,(x_n)\}</math> | |||
==''' Classification - November 24, 2011'''== | |||
===Classification === | |||
Classification is predicting a discrete random variable Y (the label) from another random variable X. It is analogous to regression, but the difference between them is that regression uses continuous values, while classification uses discrete values (labels). | |||
Consider iid data <math>\displaystyle (X_1,Y_1),(X_2,Y_2),...,(X_n,Y_n) </math> where <br> | |||
<math> X_i = (X_{i1},...,X_{id}) \in \mathbb{R}^{d} </math>, representing an object, <br> | |||
<math>\ Y_i \in Y</math> is the label of the i-th object, and <math>\ Y</math> is some finite set. | |||
We wish to determine a function <math>\ h : \mathbb{R}^{d} \rightarrow Y</math> that can predict the value of <math>\ Y_i</math> given the value <math>\ X_i</math>. When we observe a new <math> \displaystyle X</math>, predict <math>\displaystyle Y</math> to be <math> \displaystyle h(X)</math>.(We use h(x)) for the following discussion). | |||
The difference between classification and clustering is that clustering do not have <math>\ Y_i</math> and it only puts <math>\ X_i</math> into different classes. | |||
====Examples==== | |||
'''Object:''' An image of a pepper' | |||
<br>The features are defined to be colour, length, diameter, and weight. | |||
[[File:pepper.jpg|200px]] | |||
<br>'''Features(X):''' (Red,6,2,3.5) | |||
<br> '''Labels(Y):''' Red Pepper | |||
<br> The objective of classification is to use the classification function for unseen data - data for which we do not know the classification. Given the features, we want to classify the object, or in other words find the label. This is the typical classification problem. | |||
<br> '''Real Life Examples''': Face Recognition - Pictures are just a points in high dimensional space; we can represent them in vectors. Sound waves can also be classified in a similar manner using Fourier Transforms. Classification is also used to find drugs which cure disease. Molecules are classified as either a good fit into the cavity of a protein, or not. | |||
<br> In machine learning, classification is also known as supervised learning. Often we separate the data set into two parts. One is called a training set and the other, a testing set. We use the training set to establish a classifier rule and use the testing set to test its effectiveness. | |||
===''' Error Rate '''=== | |||
'''Definition''': The true error rate of a classifier h is <math> L(h) = P(h(X) \neq Y) </math> and the empirical error rate or training error rate is: | |||
<br><math> \hat{L}_h = \frac{1}{n} \sum_{i=1}^{n}I(h(X_i)\neq Y_i) </math> | |||
Where <math>\!I</math> is the indicator function. That is, <math>\!I( ) = 1</math> if the statement inside the bracket is true, and <math>\!I( ) = 0</math> otherwise. | |||
<br> The empirical error rate is the the proportion of points that have not been classified correctly. It can be shown that this estimation always underestimates the true error rate, although we do not cover it in this course. | |||
A way to get a better estimate of the error is to construct the classifier with half of the given data and use the other half to calculate the error rate. | |||
===''' Bayesians vs Frequentists'''=== | |||
Bayesians view probability as the measure of confidence that a person holds in a proposition given certain information. They state a "prior probability" exist, which represents the possibility of a event's occurrence given no information. As we add new information regarding this event, our belief adjusts according to the new information, which gives us a posterior probability. Frequentists interpret probability as a "propensity" of some event. | |||
===''' Bayes Classifier '''=== | |||
Consider the special case where <math> y \in {0,1} </math>, then | |||
<br> <math> r(x) = P(Y=1|X=x) = \frac{P(X=x|Y=1)P(Y=1)}{P(X=x)} = \frac{P(X=x|Y=1)P(Y=1)}{P(X=x|Y=1)P(Y=1)+P(X=x|Y=0)P(Y=0)} </math> | |||
<br> '''Definition''': The Bayes classification rule h* is: | |||
<br> <math>h^*(x)=\left\{\begin{matrix}1 & \mathrm{if}\ \hat{r}(x) > 0.5 \\ 0 & otherwise\end{matrix}\right.</math> | |||
<br> The set <math>\displaystyle D(h) = \{x: P(Y=1|X=x)=P(Y=0|X=x)\} </math> is called the decision boundary. | |||
<br> <math>h^*(x)=\left\{\begin{matrix}1 & \mathrm{if}\ P(Y=1|X=x) > P(Y=0|X=x) \\ 0 & otherwise\end{matrix}\right.</math> | |||
<br> '''Theorem:''' The Bayes rule is optimal, that is, if <math>\ h</math> is any other classification rule, then <math> L(h^*) \leq L(h) </math> | |||
<br> So if Bayes rule is optimal, why do we need any other method? Well, we don't always know the distributions of the data. The Bayes rule depends on unknown quantities, so we need to use the data to find some approximation to the Bayes rule. | |||
===''' Three Main Approaches '''=== | |||
1. Empirical Risk Minimization: Choose a set of classifiers H and find a <math> h^* \in H </math> that minimizes the expected value of some loss function <math>\ L(h) </math>. The distribution of (X,Y) is not known, therefore <math>\ E[L(h)]</math> must be estimated using empirical data. For more conceptual knowledge on how this approach works, please refer to http://en.wikipedia.org/wiki/Empirical_risk_minimization | |||
2. Regression: Find an estimate <math> \hat{r}(x) </math> of the function <math>\displaystyle r </math> and define: | |||
<br> <math> h^*(x)=\left\{\begin{matrix}1 & \mathrm{if}\ \hat{r}(x) > 0.5 \\ 0 & otherwise\end{matrix}\right.</math> | |||
3. Density Estimation: Estimate <math>\displaystyle P(X=x|Y=0) </math> from the <math>\displaystyle X_i's</math> for which <math>\displaystyle Y_i=0</math>, and estimate <math>\displaystyle P(X=x|Y=1)</math> from the <math>\displaystyle X_i's</math> for which <math>\displaystyle Y_i=1</math> and let: | |||
<br> <math>\displaystyle P(Y=1)=\frac{1}{n} \sum_{i=1}^{n}Y_i </math> | |||
<br> Define | |||
<br><math> \hat{r}(x)=\hat{P}(Y=1|X=x) </math> | |||
<br> and define | |||
<br><math> h(x)=\left\{\begin{matrix}1 & \mathrm{if}\ \hat{r}(x) > 0.5 \\ 0 & otherwise\end{matrix}\right.</math> | |||
<br> So we have | |||
<br><math> P(Y=1|X=x) = \frac{P(X=x|Y=1)P(Y=1)}{P(X)} </math> | |||
<br> where | |||
<br> <math>\displaystyle P(X) = P(X=x|Y=1)P(Y=1) + P(X=x|Y=0)P(Y=0) </math> | |||
===''' Multi-class Classification '''=== | |||
We want to generalize to the case that Y takes on more than two values | |||
<br> '''Theorem:''' Suppose that <math> Y \in y = \{1,...,k\} </math>, the optimal rule is | |||
<br> <math>\displaystyle h^*(x) = {\operatorname{arg\,max}}_k\{P(Y=k|X=x)\} </math> | |||
<br> where | |||
<br> <math> P(Y=k|X=x)=\frac{f_k(x)\pi_k}{\sum_{k}f_k(x)\pi_k} </math> | |||
<br> with | |||
<br> <math>\displaystyle f_k(x) = P(X=x|Y=k)</math> | |||
<br> <math> \displaystyle \pi_k = P(Y=k) </math> | |||
A direct interpretation of this method called "naive Bayes classifier", is that the classifier chooses an y value which maximizes the probability of getting the current x value. In other words, the value y here can be viewed as a parameter as in maximum likelihood estimation, where the optimal value is <math>\displaystyle y^* = {\operatorname{arg\,max}}\{f(y|x)\} </math>. | |||
==''' LDA, QDA and FDA - November 29, 2011'''== | |||
===''' Linear Discriminant Analysis (LDA) and Quadratic Discriminant Analysis (QDA)'''=== | |||
The simplest approach to classification is to use the third approach listed above and assume a parametric model for densities. | |||
Applying Bayes rule: | |||
<br> <math> P(Y=k|X=x) = \frac{P(X=x|Y=k)P(Y=k)}{P(X=x)} = \frac{P(X=x|Y=k)P(Y=k)}{\sum_{i}P(X=x|Y=i)P(Y=i)} </math> <br> | |||
<br> | |||
We notice that the denominator value is always the same independent of k; thus will be cancelled out. Hence we will only need to evaluate the numerator value. | |||
Define class conditional distribution, prior distribution and posterior distribution as follows: | |||
<br> <math>\ f_k(x)=P(X=x|Y=k),{\mathbf\pi_k}=P(Y=k), P(Y=k|X=x) </math> <br> | |||
<br> | |||
'''LDA (Linear Discriminant Analysis)''' <br> | |||
LDA is a classifier which makes the 2 assumptions: | |||
<br> | |||
* The class conditional distribution is Gaussian | |||
::<math>\ f_k(x)= \frac{1}{(2\pi)^{d/2}|\Sigma_k|^{1/2}} | |||
\exp\left(-\frac{1}{2}({\mathbf x}-{\mathbf\mu_k})^T{\mathbf\Sigma_k}^{-1}({\mathbf x}-{\mathbf\mu_k}) | |||
\right),x\in \mathbb{R}^d</math> | |||
<br> | |||
where <math>|\mathbf\Sigma_k|</math> is the determinant of <math>\mathbf\Sigma_k</math> | |||
* The two classes share the same covariance matrix | |||
::<math> {\mathbf\Sigma}_0={\mathbf\Sigma}_1={\mathbf\Sigma} </math> | |||
<br> | |||
Thus the class conditional distribution for <math>k</math> in LDA is as follows: | |||
<br> <math>\ f_k(x)= \frac{1}{(2\pi)^{d/2}|\Sigma|^{1/2}} | |||
\exp\left(-\frac{1}{2}({\mathbf x}-{\mathbf\mu_k})^T{\mathbf\Sigma}^{-1}({\mathbf x}-{\mathbf\mu_k}) | |||
\right),x\in \mathbb{R}^d</math><br> | |||
To demonstrate, suppose we have a two class scenario, i.e <math> y \in \{0,1 \} </math>, and we want to compute the decision boundary for this LDA. Recall that the decision boundary is a set of points such that: | |||
<br> <math>\ P(Y=0|X=x)=P(Y=1|X=x) </math><br> | |||
where the probability is given by: | |||
<br> <math> P(Y=0|X=x) = \frac{f_0(x){\mathbf\pi_0}}{\sum_{k=0}^{1}f_k(x){\mathbf\pi_k}},P(Y=1|X=x) = \frac{f_1(x){\mathbf\pi_1}}{\sum_{k=0}^{1}f_k(x){\mathbf\pi_k}} </math> <br> | |||
Hence we want to find <math> {\mathbf x} </math> such that | |||
<br> <math>\ P(Y=0|X=x)=P(Y=1|X=x) </math><br> | |||
<br> <math> \frac{f_0(x){\mathbf\pi_0}}{\sum_{k=0}^{1}f_k(x){\mathbf\pi_k}} = \frac{f_1(x){\mathbf\pi_1}}{\sum_{k=0}^{1}f_k(x){\mathbf\pi_k}} </math> <br> | |||
<br> <math> f_0(x){\mathbf\pi_0}=f_1(x){\mathbf\pi_1} </math> <br> | |||
<br> | |||
Upon expanding <math>\ f_k(x) </math>, we get | |||
<br><math> | |||
\frac{1}{(2\pi)^{d/2}|\Sigma|^{1/2}} | |||
\exp\left(-\frac{1}{2}({\mathbf x}-{\mathbf\mu_0})^T{\mathbf\Sigma}^{-1}({\mathbf x}-{\mathbf\mu_0}) | |||
\right) * \pi_0 | |||
</math> | |||
<math> | |||
=\frac{1}{(2\pi)^{d/2}|\Sigma|^{1/2}} | |||
\exp\left(-\frac{1}{2}({\mathbf x}-{\mathbf\mu_1})^T{\mathbf\Sigma}^{-1}({\mathbf x}-{\mathbf\mu_1}) | |||
\right) * \pi_1 | |||
</math><br> | |||
<br> | |||
Rearranging the equation and taking logarithm on both sides we get: | |||
<br><math> | |||
-\frac{1}{2}({\mathbf x}-{\mathbf\mu_0})^T{\mathbf\Sigma}^{-1}({\mathbf x}-{\mathbf\mu_0})+\log({\mathbf\pi_0}) | |||
=-\frac{1}{2}({\mathbf x}-{\mathbf\mu_1})^T{\mathbf\Sigma}^{-1}({\mathbf x}-{\mathbf\mu_1})+\log({\mathbf\pi_1}) | |||
</math><br> | |||
That is, after expanding the quadratic form of our vector values, | |||
<br><math> | |||
\frac{1}{2} | |||
\left | |||
[-{\mathbf x}^T{\mathbf\Sigma}^{-1}{\mathbf x} | |||
-{\mathbf\mu_1}^T{\mathbf\Sigma}^{-1}{\mathbf\mu_1} | |||
+2{\mathbf x}^T{\mathbf\Sigma}^{-1}{\mathbf\mu_1} | |||
+{\mathbf x}^T{\mathbf\Sigma}^{-1}{\mathbf x} | |||
+{\mathbf\mu_0}^T{\mathbf\Sigma}^{-1}{\mathbf\mu_0} | |||
-2{\mathbf x}^T{\mathbf\Sigma}^{-1}{\mathbf\mu_0} | |||
\right] | |||
+\log(\frac{\mathbf\pi_1}{\mathbf\pi_0}) | |||
=0 | |||
</math><br> | |||
Cancelling out the terms and notice that <math> {\mathbf\mu_k}^T{\mathbf\Sigma}^{-1}{\mathbf\mu_k} </math> is constant and <math> {\mathbf x}^T{\mathbf\Sigma}^{-1}{\mathbf\mu_k} </math> a linear expression of <math>\ x </math>, the final equation is simply linear system of equations: | |||
<br> <math> {\mathbf x}^T{\mathbf\Sigma}^{-1}({\mathbf\mu_1}-{\mathbf\mu_0}) | |||
+\frac{1}{2}\left(-{\mathbf\mu_1}^T{\mathbf\Sigma}^{-1}{\mathbf\mu_1} | |||
+{\mathbf\mu_0}^T{\mathbf\Sigma}^{-1}{\mathbf\mu_0}\right) | |||
+\log(\frac{\mathbf\pi_1}{\mathbf\pi_0}) | |||
=0 | |||
</math> | |||
'''QDA (Quadratic Discriminant Analysis)''' | |||
If we relax the assumption in LDA that the covariances are identical between classes, the decision boundary becomes quadratic and we get QDA. Mathematically we mean that <math> {\mathbf\Sigma}_0\ne{\mathbf\Sigma}_1 </math>. In fact, we may also drop the assumption of a bi-class problem. QDA is therefore a more generalized version of LDA. | |||
The general form of the boundary for a K-class problem is | |||
<math>\delta_k (x) = -\frac{1}{2}ln|{\mathbf\Sigma}_k| -\frac{1}{2}(x-{\mathbf\mu_k})^T{\mathbf\Sigma_k}^{-1} (x-{\mathbf\mu_k}) + ln({\mathbf\pi_k})</math> | |||
and the classifier will be | |||
<br><math>\ h(x) = \underset{k}{\operatorname{arg\,max}} \, (\delta_k) </math><br> | |||
In other words we would assign each <math>\ x </math> to the label <math>\ k </math> that produces the largest <math>\ \delta_k (x)</math>. | |||
The reason for this becomes more obvious if we consider the components of <math>\ \delta_k (x)</math>. | |||
Notice that the middle component (without the coefficient), <math>\ (x-{\mathbf\mu_k})^T{\mathbf\Sigma_k}^{-1} (x-{\mathbf\mu_k}) </math>, is expressing the squared Mahalanobis distance between <math>\ x </math> and <math>\ {\mathbf\mu_k} </math>, the mean of class <math>\ k </math>. Therefore, by requiring that <math>\ \delta_k (x)</math> is maximized, we are requiring this distance to be minimized. This makes sense from a classification perspective since it is intuitive to label <math>\ x </math> with the class that it is closest to (on an average basis). | |||
Also, to illustrate the role that <math>\ ln({\mathbf\pi_k})</math> plays, consider the case where each class is comprised of the same number of points so that <math>\ ln({\mathbf\pi_i})\ =\ ln({\mathbf\pi_j})\ \forall i,j \in y={1,...,K}</math>. Then this term makes no difference in determining the appropriate label. But if a particular class <math>\ k </math> is comprised of many more points than others, then <math>\ ln({\mathbf\pi_k})</math> is larger and increases the probability for this class (label) being assigned. This is intuitive as <math>\ \pi_k </math> represents the probability of a class being <math>\ k </math>. Therefore it must contribute to the probability that a point is in the class. | |||
In practice when we estimate the parameters, | |||
<br><math> \hat{\mathbf\pi_k}=\frac{n_k}{n},\hat{\mathbf\mu_k}=\frac{1}{n_k}\sum_{i:y_i=k}{x_i}</math><br> | |||
<br><math> \hat{\mathbf\Sigma}_k=\frac{1}{n_k}\sum_{i:y_i=k}({\mathbf x}_i-{\mathbf\mu_k})({\mathbf x}_i-{\mathbf\mu_k})^T</math><br> | |||
<br> | |||
If we assume <math> {\mathbf\Sigma}_k={\mathbf\Sigma} </math> for all <math>\ k </math>, then | |||
<br><math> \hat{\mathbf\Sigma}=\frac{\sum_{r=1}^{k}n_r{\mathbf\Sigma_r}}{n}</math><br> | |||
which is the weighted average of all <math>{\mathbf\Sigma_k}</math>. | |||
===''' Fischer Discriminant Analysis (optional) '''=== | |||
Recall that PCA finds the direction of maximum variance. Imagine PCA, MDS or Isomap as a pre-step to classification as they all reduce the dimension of the data. But if the classification is not based on the maximum variance, PCA will not work well in this case. | |||
Suppose we want to reduce a set of two-dimensional and two-class data to one dimension in order to classify them. One way to achieve this is to draw the data set in each class as close as possible (almost collapse into one point) and set these classes far apart to distinguish them. This is the intuitive interpretation of FDA. FDA does not carry the assumption of QDA and LDA, where the conditional probability distributions are a normally distributed, this allows a more flexible approach to classifying a data set. | |||
We want to find vector <math>\!\omega</math> to project every point <math>\!x</math> to <math>\!{{\omega }^{T}}x</math> so that maximum variation occurs between classes. | |||
Say there are two classes 0 and 1. | |||
The points that label 0 has a mean of <math>\!{{\mu }_{0}}</math> and the points that label 1 has a mean of <math>\!{{\mu }_{1}}</math>. | |||
Then <math>{{\mu }_{0}}\to {{\omega }^{T}}{{\mu }_{0}}</math>, and | |||
<math>{{\mu }_{1}}\to {{\omega }^{T}}{{\mu }_{1}}</math> | |||
To make the two classes as far away as possible, we want to | |||
<math>\underset{\omega }{\mathop{\max }}\,||{{\omega }^{T}}{{\mu }_{0}}-{{\omega }^{T}}{{\mu }_{1}}||^2</math> | |||
In other words, | |||
<math>\underset{\omega }{\mathop{\max }}\,{{\omega }^{T}}({{\mu }_{0}}-{{\mu }_{1}}){{({{\mu }_{0}}-{{\mu }_{1}})}^{T}}\omega </math> | |||
We define <math>{{S}_{B}}:=({{\mu }_{0}}-{{\mu }_{1}}){{({{\mu }_{0}}-{{\mu }_{1}})}^{T}}</math>, which is the between-class covariance. | |||
On the other hand, we want the variance within the same class as small as possible, we want to | |||
<math>\underset{w}{\mathop{\min }}\,{{\omega }^{T}}{{\sum }_{0}}\omega +{{\omega }^{T}}{{\sum }_{1}}\omega </math> | |||
In other words, | |||
<math>\underset{w}{\mathop{\min }}\,{{\omega }^{T}}({{\sum }_{0}}+{{\sum }_{1}})\omega </math> | |||
We define <math>{{S}_{W}}={{\sum }_{0}}+{{\sum }_{1}}</math>, the within-class covariance. | |||
Our question becomes, | |||
<math>\underset{w}{\mathop{\max }}\,=\frac{{{\omega }^{T}}{{S}_{B}}\omega }{{{\omega }^{T}}{{S}_{W}}\omega }</math> | |||
Or equivalently, | |||
<math>\underset{{}}{\mathop{\max }}\,{{\omega }^{T}}{{S}_{B}}\omega </math> such that <math>\!{{\omega }^{T}}{{S}_{W}}\omega =1</math> | |||
Using the langrage multiplier, <math>\!L(\omega ,\lambda )={{\omega }^{T}}{{S}_{B}}\omega -\lambda ({{\omega }^{T}}{{S}_{W}}\omega -1)</math>, we have | |||
<math>\frac{\partial L}{\partial \omega }=0</math> | |||
<math>\Rightarrow {{S}_{B}}\omega =\lambda {{S}_{W}}\omega </math> | |||
<math>\Rightarrow S_{W}^{-1}{{S}_{B}}\omega =\lambda \omega </math> | |||
>> | Therefore, <math>\!\omega</math> is the eigenvector of <math>S_{W}^{-1}{{S}_{B}}</math> (the eigenvector of the largest <math>\lambda </math>) | ||
Generally, FDA is a better choice than PCA in classification. | |||
Latest revision as of 09:45, 30 August 2017
Please contribute to the discussion of splitting up this page into multiple pages on the talk page.
Editor Sign Up
Notation
The following guidelines on notation were posted on the Wiki Course Note page for STAT 946. Add to them as necessary for consistent notation on this page.
Capital letters will be used to denote random variables and lower case letters denote observations for those random variables:
- [math]\displaystyle{ \{X_1,\ X_2,\ \dots,\ X_n\} }[/math] random variables
- [math]\displaystyle{ \{x_1,\ x_2,\ \dots,\ x_n\} }[/math] observations of the random variables
The joint probability mass function can be written as:
or as shorthand, we can write this as [math]\displaystyle{ p( x_1, x_2, \dots, x_n ) }[/math]. In these notes both types of notation will be used. We can also define a set of random variables [math]\displaystyle{ X_Q }[/math] where [math]\displaystyle{ Q }[/math] represents a set of subscripts.
Sampling - September 20, 2011
The meaning of sampling is to generate data points or numbers such that these data follow a certain distribution.
i.e. From [math]\displaystyle{ x \sim~f(x) }[/math] sample [math]\displaystyle{ \,x_{1}, x_{2}, ..., x_{1000} }[/math]
In practice, it maybe difficult to find the joint distribution of random variables. We will explore different methods for simulating random variables, and how to draw conclusions using the simulated data.
Sampling from Uniform Distribution
Computers cannot generate random numbers as they are deterministic; however they can produce pseudo random numbers using algorithms. Generated numbers mimic the properties of random numbers but they are never truly random. One famous algorithm that is considered highly reliable is the Mersenne twister[1], which generates random numbers in an almost uniform distribution.
Multiplicative Congruential
- involves four parameters: integers [math]\displaystyle{ \,a, b, m }[/math], and an initial value [math]\displaystyle{ \,x_0 }[/math] which we call the seed
- a sequence of integers is defined as
- [math]\displaystyle{ x_{k+1} \equiv (ax_{k} + b) \mod{m} }[/math]
Example: [math]\displaystyle{ \,a=13, b=0, m=31, x_0=1 }[/math] creates a uniform histogram.
MATLAB code for generating 1000 random numbers using the multiplicative congruential method:
a = 13;
b = 0;
m = 31;
x(1) = 1;
for ii = 2:1000
x(ii) = mod(a*x(ii-1)+b, m);
end
MATLAB code for displaying the values of x generated:
x
MATLAB code for plotting the histogram of x:
hist(x)
Histogram Output:
Facts about this algorithm:
- In this example, the first 30 terms in the sequence are a permutation of integers from 1 to 30 and then the sequence repeats itself. In the general case, this algorithm has a period of m-1.
- Values are between 0 and m-1, inclusive.
- Dividing the numbers by m-1 yields numbers in the interval [0,1].
- MATLAB's
randfunction once used this algorithm with a= 75, b= 0, m= 231-1,for reasons described in Park and Miller's 1988 paper "Random Number Generators: Good Ones are Hard to Find" (available online). - Visual Basic's
RNDfunction also used this algorithm with a= 1140671485, b= 12820163, m= 224. (Reference)
Inverse Transform Method
This is a basic method for sampling. Theoretically using this method we can generate sample numbers at random from any probability distribution once we know its cumulative distribution function (cdf). This method is very efficient computationally if the cdf of can be analytically inverted.
Theorem
Take [math]\displaystyle{ U \sim~ \mathrm{Unif}[0, 1] }[/math] and let [math]\displaystyle{ \ X = F^{-1}(U) }[/math]. Then [math]\displaystyle{ \ X }[/math] has a cumulative distribution function of [math]\displaystyle{ F(\cdot) }[/math], ie. [math]\displaystyle{ F(x)=P(X \leq x) }[/math], where [math]\displaystyle{ F^{-1}(\cdot) }[/math] is the inverse of [math]\displaystyle{ F(\cdot) }[/math].
Proof
Recall that
- [math]\displaystyle{ P(a \leq X\lt b)=\int_a^{b} f(x) dx }[/math]
- [math]\displaystyle{ cdf=F(x)=P(X \leq x)=\int_{-\infty}^{x} f(x) dx }[/math]
Note that if [math]\displaystyle{ U \sim~ \mathrm{Unif}[0, 1] }[/math], we have [math]\displaystyle{ P(U \leq a)=a }[/math]
- [math]\displaystyle{ \begin{align} P(F^{-1}(U) \leq x) &{}= P(F(F^{-1}(U)) \leq F(x)) \\ &{}= P(U \leq F(x)) \\ &{}= F(x) \end{align} }[/math]
Completing the proof.
Continuous Case
Generally it takes two steps to get random numbers using this method.
- Step 1. Draw [math]\displaystyle{ U \sim~ \mathrm{Unif}[0, 1] }[/math]
- Step 2. X=F −1(U)
Example
Take the exponential distribution for example
- [math]\displaystyle{ \,f(x)={\lambda}e^{-{\lambda}x} }[/math]
- [math]\displaystyle{ \,F(x)=\int_0^x {\lambda}e^{-{\lambda}u} du=[-e^{-{\lambda}u}]_0^x=1-e^{-{\lambda}x} }[/math]
Let: [math]\displaystyle{ \,F(x)=y }[/math]
- [math]\displaystyle{ \,y=1-e^{-{\lambda}x} }[/math]
- [math]\displaystyle{ \,ln(1-y)={-{\lambda}x} }[/math]
- [math]\displaystyle{ \,x=\frac{ln(1-y)}{-\lambda} }[/math]
- [math]\displaystyle{ \,F^{-1}(x)=\frac{-ln(1-x)}{\lambda} }[/math]
Therefore, to get a exponential distribution from a uniform distribution takes 2 steps.
- Step 1. Draw [math]\displaystyle{ U \sim~ \mathrm{Unif}[0, 1] }[/math]
- Step 2. [math]\displaystyle{ x=\frac{-ln(1-U)}{\lambda} }[/math]
Note: If U~Unif[0, 1], then (1 - U) and U have the same distribution. This allows us to slightly simplify step 2 into an alternate form:
- Alternate Step 2. [math]\displaystyle{ x=\frac{-ln(U)}{\lambda} }[/math]
MATLAB code
for exponential distribution case,assuming [math]\displaystyle{ \lambda=0.5 }[/math]
for ii = 1:1000
u = rand;
x(ii) = -log(1-u)/0.5;
end
hist(x)
MATLAB result
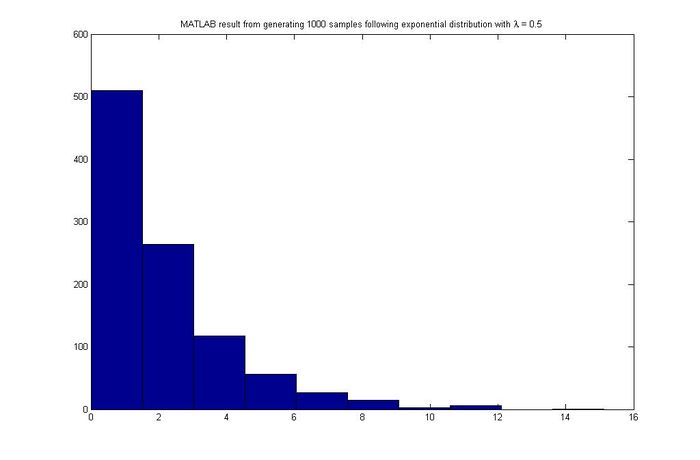
Discrete Case - September 22, 2011
This same technique can be applied to the discrete case. Generate a discrete random variable [math]\displaystyle{ \,x }[/math] that has probability mass function [math]\displaystyle{ \,P(X=x_i)=P_i }[/math] where [math]\displaystyle{ \,x_0\lt x_1\lt x_2... }[/math] and [math]\displaystyle{ \,\sum_i P_i=1 }[/math]
- Step 1. Draw [math]\displaystyle{ u \sim~ \mathrm{Unif}[0, 1] }[/math]
- Step 2. [math]\displaystyle{ \,x=x_i }[/math] if [math]\displaystyle{ \,F(x_{i-1})\lt u \leq F(x_i) }[/math]
Example
Let x be a discrete random variable with the following probability mass function:
- [math]\displaystyle{ \begin{align} P(X=0) = 0.3 \\ P(X=1) = 0.2 \\ P(X=2) = 0.5 \end{align} }[/math]
Given the pmf, we now need to find the cdf.
We have:
- [math]\displaystyle{ F(x) = \begin{cases} 0 & x \lt 0 \\ 0.3 & x \leq 0 \\ 0.5 & x \leq 1 \\ 1 & x \leq 2 \end{cases} }[/math]
We can apply the inverse transform method to obtain our random numbers from this distribution.
Pseudo Code for generating the random numbers:
Draw U ~ Unif[0,1] if U <= 0.3 return 0 else if 0.3 < U <= 0.5 return 1 else if 0.5 < U <= 1 return 2
MATLAB code for generating 1000 random numbers in the discrete case:
for ii = 1:1000
u = rand;
if u <= 0.3
x(ii) = 0;
else if u <= 0.5
x(ii) = 1;
else
x(ii) = 2;
end
end
Matlab Output:
Pseudo code for the Discrete Case:
1. Draw U ~ Unif [0,1]
2. If [math]\displaystyle{ U \leq P_0 }[/math], deliver X= x0
3. Else if [math]\displaystyle{ U \leq P_0 + P_1 }[/math], deliver X= x1
4. Else If [math]\displaystyle{ U \leq P_0 +....+ P_k }[/math], deliver X= xk
Limitations
This method is useful, but it's not practical in many cases because we can't always obtain [math]\displaystyle{ F }[/math] or [math]\displaystyle{ F^{-1} }[/math] (some functions are not integrable or invertible), and sometimes [math]\displaystyle{ f(x) }[/math] cannot be obtained in closed form. Let's look at some examples:
- Continuous case
If we want to use this method to draw the pdf of normal distribution, we may find ourselves geting stuck when trying to find its cdf. The simplest case of normal distribution is [math]\displaystyle{ f(x)=\frac{1}{\sqrt{2\pi}}e^{-\frac{x^2}{2}} }[/math], whose cdf is [math]\displaystyle{ F(x)=\frac{1}{\sqrt{2\pi}}\int_{-\infty}^{x}{e^{-\frac{u^2}{2}}}du }[/math]. This integral cannot be expressed in terms of elementary functions. So evaluating it and then finding the inverse is a very difficult task.
- Discrete case
It is easy for us to simulate when there are only a few values taken by the particular random variable, like the case above. And it is easy to simulate the binomial distribution [math]\displaystyle{ X \sim~ \mathrm{B}(n,p) }[/math] when the parameter n is not too large. But when n takes on values that are very large, say 50, it is hard to do so.
Acceptance/Rejection Method
The aforementioned difficulties of the inverse transform method motivates a sampling method that does not require analytically calculating cdf's and their inverses, which is the acceptance/rejection sampling method. Here, [math]\displaystyle{ \displaystyle f(x) }[/math] is approximated by another function, say [math]\displaystyle{ \displaystyle g(x) }[/math], with the idea being that [math]\displaystyle{ \displaystyle g(x) }[/math] is a "nicer" function to work with than [math]\displaystyle{ \displaystyle f(x) }[/math].
Suppose we assume the following:
1. There exists another distribution [math]\displaystyle{ \displaystyle g(x) }[/math] that is easier to work with and that you know how to sample from, and
2. There exists a constant c such that [math]\displaystyle{ f(x) \leq c \cdot g(x) }[/math] for all x
Under these assumptions, we can sample from [math]\displaystyle{ \displaystyle f(x) }[/math] by sampling from [math]\displaystyle{ \displaystyle g(x) }[/math]
General Idea
Looking at the image below we have graphed [math]\displaystyle{ c \cdot g(x) }[/math] and [math]\displaystyle{ \displaystyle f(x) }[/math].
Using the acceptance/rejection method we will accept some of the points from [math]\displaystyle{ \displaystyle g(x) }[/math] and reject some of the points from [math]\displaystyle{ \displaystyle g(x) }[/math]. The points that will be accepted from [math]\displaystyle{ \displaystyle g(x) }[/math] will have a distribution similar to [math]\displaystyle{ \displaystyle f(x) }[/math]. We can see from the image that the values around [math]\displaystyle{ \displaystyle x_1 }[/math] will be sampled more often under [math]\displaystyle{ c \cdot g(x) }[/math] than under [math]\displaystyle{ \displaystyle f(x) }[/math], so we will have to reject more samples taken at x1. Around [math]\displaystyle{ \displaystyle x_2 }[/math] the number of samples that are drawn and the number of samples we need are much closer, so we accept more samples that we get at [math]\displaystyle{ \displaystyle x_2 }[/math]. We should remember to take these considerations into account when evaluating the efficiency of a given acceptance-rejection method. Rejecting a high proportion of samples ultimately leaves us with a longer time until we retrieve our desired distribution. We should question whether a better function [math]\displaystyle{ g(x) }[/math] can be chosen in our situation.
Procedure
1. Draw y ~ g
2. Draw U ~ Unif [0,1]
3. If [math]\displaystyle{ U \leq \frac{f(y)}{c \cdot g(y)} }[/math] then x=y; else return to 1
Note that the choice of [math]\displaystyle{ c }[/math] plays an important role in the efficiency of the algorithm. We want [math]\displaystyle{ c \cdot g(x) }[/math] to be "tightly fit" over [math]\displaystyle{ f(x) }[/math] to increase the probability of accepting points, and therefore reducing the number of sampling attempts. Mathematically, we want to minimize [math]\displaystyle{ c }[/math] such that [math]\displaystyle{ f(x) \leq c \cdot g(x) \ \forall x }[/math]. We do this by setting
[math]\displaystyle{ \frac{d}{dx}(\frac{f(x)}{g(x)}) = 0 }[/math], solving for a maximum point [math]\displaystyle{ x_0 }[/math] and setting [math]\displaystyle{ c = \frac{f(x_0)}{g(x_0)}. }[/math]
Proof
Mathematically, we need to show that the sample points given that they are accepted have a distribution of f(x).
[math]\displaystyle{ \begin{align} P(y|accepted) &= \frac{P(y, accepted)}{P(accepted)} \\ &= \frac{P(accepted|y) P(y)}{P(accepted)}\end{align} }[/math] (Bayes' Rule)
[math]\displaystyle{ \displaystyle P(y) = g(y) }[/math]
[math]\displaystyle{ P(accepted|y) =P(u\leq \frac{f(y)}{c \cdot g(y)}) =\frac{f(y)}{c \cdot g(y)} }[/math],where u ~ Unif [0,1]
[math]\displaystyle{ P(accepted) = \sum P(accepted|y)\cdot P(y)=\int^{}_y \frac{f(s)}{c \cdot g(s)}g(s) ds=\int^{}_y \frac{f(s)}{c} ds=\frac{1}{c} \cdot\int^{}_y f(s) ds=\frac{1}{c} }[/math]
So,
[math]\displaystyle{ P(y|accepted) = \frac{ \frac {f(y)}{c \cdot g(y)} \cdot g(y)}{\frac{1}{c}} =f(y) }[/math]
Continuous Case
Example
Sample from Beta(2,1)
In general:
[math]\displaystyle{ Beta(\alpha, \beta) = \frac{\Gamma (\alpha + \beta)}{\Gamma(\alpha)\Gamma(\beta)} }[/math] [math]\displaystyle{ \displaystyle x^{\alpha-1} }[/math] [math]\displaystyle{ \displaystyle(1-x)^{\beta-1} }[/math], [math]\displaystyle{ \displaystyle 0\lt x\lt 1 }[/math]
Note: [math]\displaystyle{ \!\Gamma(n) = (n-1)! }[/math] if n is a positive integer
[math]\displaystyle{ \begin{align} f(x) &= Beta(2,1) \\ &= \frac{\Gamma(3)}{\Gamma(2)\Gamma(1)} x^1(1-x)^0 \\ &= \frac{2!}{1! 0!}\cdot (1) x \\ &= 2x \end{align} }[/math]
We want to choose [math]\displaystyle{ \displaystyle g(x) }[/math] that is easy to sample from. So we choose [math]\displaystyle{ \displaystyle g(x) }[/math] to be uniform distribution on [math]\displaystyle{ \ (0,1) }[/math] since that is the domain for the Beta function.
We now want a constant c such that [math]\displaystyle{ f(x) \leq c \cdot g(x) }[/math] for all x from Unif(0,1)
So,
[math]\displaystyle{ c \geq \frac{f(x)}{g(x)} }[/math], for all x from (0,1)
[math]\displaystyle{ \begin{align}c &\geq max (\frac {f(x)}{g(x)}, 0\lt x\lt 1) \\
&= max (\frac {2x}{1},0\lt x\lt 1) \\
&= 2 \end{align} }[/math]
Now that we have c =2,
1. Draw y ~ g(x) => Draw y ~ Unif [0,1]
2. Draw u ~ Unif [0,1]
3. if [math]\displaystyle{ u \leq \frac{2y}{2 \cdot 1} }[/math] then x=y; else return to 1
MATLAB code for generating 1000 samples following Beta(2,1):
close all
clear all
ii=1;
while ii < 1000
y = rand;
u = rand;
if u <= y
x(ii)=y;
ii=ii+1;
end
end
hist(x)
MATLAB result
Discrete Example
Generate random variables according to the p.m.f:
- [math]\displaystyle{ \begin{align} P(Y=1) = 0.15 \\ P(Y=2) = 0.22 \\ P(Y=3) = 0.33 \\ P(Y=4) = 0.10 \\ P(Y=5) = 0.20 \end{align} }[/math]
find a g(y) discrete uniform distribution from 1 to 5
[math]\displaystyle{ c \geq \frac{P(y)}{g(y)} }[/math]
[math]\displaystyle{ c = \max \left(\frac{P(y)}{g(y)} \right) }[/math]
[math]\displaystyle{ c = \max \left(\frac{0.33}{0.2} \right) = 1.65 }[/math] Since P(Y=3) is the max of P(Y) and g(y) = 0.2 for all y.
1. Generate Y according to the discrete uniform between 1 - 5
2. U ~ unif[0,1]
3. If [math]\displaystyle{ U \leq \frac{P(y)}{1.65 \times 0.2} \leq \frac{P(y)}{0.33} }[/math], then x = y; else return to 1.
In MATLAB, the code would be:
py = [0.15 0.22 0.33 0.1 0.2];
ii =1;
while ii <= 1000
y = unidrnd(5);
u = rand;
if u <= py(y)/0.33
x(ii) = y;
ii = ii+1;
end
end
hist(x);
MATLAB result
Limitations
Most of the time we have to sample many more points from g(x) before we can obtain an acceptable amount of samples from f(x), hence this method may not be computationally efficient. It depends on our choice of g(x). For example, in the example above to sample from Beta(2,1), we need roughly 2000 samples from g(X) to get 1000 acceptable samples of f(x).
In addition, in situations where a g(x) function is chosen and used, there can be a discrepancy between the functional behaviors of f(x) and g(x) that render this method unreliable. For example, given the normal distribution function as g(x) and a function of f(x) with a "fat" mid-section and "thin tails", this method becomes useless as more points near the two ends of f(x) will be rejected, resulting in a tedious and overwhelming number of sampling points having to be sampled due to the high rejection rate of such a method.
Sampling From Gamma and Normal Distribution - September 27, 2011
Sampling From Gamma
Gamma Distribution
The Gamma function is written as [math]\displaystyle{ X \sim~ Gamma (t, \lambda) }[/math]
- [math]\displaystyle{ F(x) = \int_{0}^{\lambda x} \frac{e^{-y}y^{t-1}}{(t-1)!} dy }[/math]
If you have t samples of the exponential distribution,
[math]\displaystyle{ \begin{align} X_1 \sim~ Exp(\lambda)\\ \vdots \\ X_t \sim~ Exp(\lambda) \end{align}
}[/math]
The sum of these t samples has a gamma distribution,
- [math]\displaystyle{ X_1+X_2+ ... + X_t \sim~ Gamma (t, \lambda) }[/math]
- [math]\displaystyle{ \sum_{i=1}^{t} X_i \sim~ Gamma (t, \lambda) }[/math] where [math]\displaystyle{ X_i \sim~Exp(\lambda) }[/math]
Method
Suppose we want to sample [math]\displaystyle{ \ k }[/math] points from [math]\displaystyle{ \ Gamma (t, \lambda) }[/math].
We can sample the exponential distribution using the inverse transform method from the previous class,
- [math]\displaystyle{ \,f(x)={\lambda}e^{-{\lambda}x} }[/math]
- [math]\displaystyle{ \,F^{-1}(u)=\frac{-ln(1-u)}{\lambda} }[/math]
- [math]\displaystyle{ \,F^{-1}(u)=\frac{-ln(u)}{\lambda} }[/math]
(1 - u) is the same as x since [math]\displaystyle{ U \sim~ unif [0,1] }[/math]
- [math]\displaystyle{ \begin{align} \frac{-ln(u_1)}{\lambda} - \frac{ln(u_2)}{\lambda} - ... - \frac{ln(u_t)}{\lambda} = x_1\\ \vdots \\ \frac{-ln(u_1)}{\lambda} - \frac{ln(u_2)}{\lambda} - ... - \frac{ln(u_t)}{\lambda} = x_k \end{align}
: }[/math]
- [math]\displaystyle{ \frac {-\sum_{i=1}^{t} ln(u_i)}{\lambda} = x }[/math]
MATLAB code for a Gamma(3,1) is
x = sum(-log(rand(1000,3)),2); hist(x)
And the Histogram of X follows a Gamma distribution with long tail:
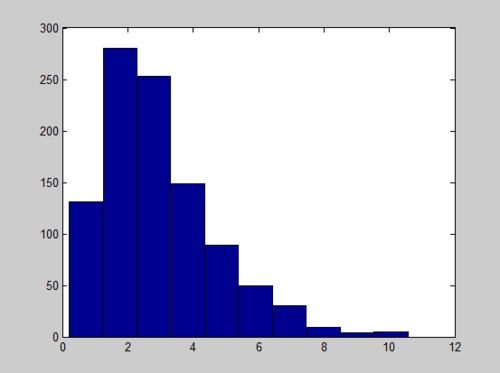
We can improve the quality of histogram by adjusting the Matlab code to include the number of bins we want: hist(x, number_of_bins)
x = sum(-log(rand(20000,3)),2); hist(x,40)
R code for a Gamma(3,1) is
a<-apply(-log(matrix(runif(3000),nrow=1000)),1,sum); hist(a);
Histogram:
Here is another histogram of Gamma coding with R
a<-apply(-log(matrix(runif(3000),nrow=1000)),1,sum); hist(a,freq=F); lines(density(a),col="blue"); rug(jitter(a));
Sampling from Normal Distribution using Box-Muller Transform - September 29, 2011
Procedure
- Generate [math]\displaystyle{ \displaystyle u_1 }[/math] and [math]\displaystyle{ \displaystyle u_2 }[/math], two values sampled from a uniform distribution between 0 and 1.
- Set [math]\displaystyle{ \displaystyle R^2 = -2log(u_1) }[/math] so that [math]\displaystyle{ \displaystyle R^2 }[/math] is exponential with mean 2
Set [math]\displaystyle{ \!\theta = 2*\pi*u_2 }[/math] so that [math]\displaystyle{ \!\theta }[/math] ~ [math]\displaystyle{ \ Unif[0, 2\pi] }[/math] - Set [math]\displaystyle{ \displaystyle X = R cos(\theta) }[/math]
Set [math]\displaystyle{ \displaystyle Y = R sin(\theta) }[/math]
Justification
Suppose we have X ~ N(0, 1) and Y ~ N(0, 1) where X and Y are independent normal random variables. The relative probability density function of these two random variables using Cartesian coordinates is:
[math]\displaystyle{ f(X, Y) dxdy= f(X) f(Y) dxdy= \frac{1}{\sqrt{2\pi}}e^{-x^2/2} \frac{1}{\sqrt{2\pi}}e^{-y^2/2} dxdy= \frac{1}{2\pi}e^{-(x^2+y^2)/2}dxdy }[/math]
In polar coordinates [math]\displaystyle{ \displaystyle R^2 = x^2 + y^2 }[/math], so the relative probability density function of these two random variables using polar coordinates is:
[math]\displaystyle{ f(R, \theta) = \frac{1}{2\pi}e^{-R^2/2} }[/math]
If we have [math]\displaystyle{ \displaystyle R^2 \sim exp(1/2) }[/math] and [math]\displaystyle{ \!\theta \sim unif[0, 2\pi] }[/math] we get an equivalent relative probability density function. Notice that when doing a two by two linear transformation, the determinant of the Jacobian needs to be included in the change of variable formula where:
[math]\displaystyle{ |J|=\left|\frac{\partial(x,y)}{\partial(R,\theta)}\right|= \left|\begin{matrix}\frac{\partial x}{\partial R}&\frac{\partial x}{\partial \theta}\\\frac{\partial y}{\partial R}&\frac{\partial y}{\partial \theta}\end{matrix}\right|=R }[/math]
[math]\displaystyle{ f(X, Y) dxdy = f(R, \theta)|J|dRd\theta = \frac{1}{2\pi}e^{-R^2/2}R dRd\theta= \frac{1}{4\pi}e^{-\frac{s}{2}} dSd\theta }[/math]
where [math]\displaystyle{ S=R^2. }[/math]
Therefore we can generate a point in polar coordinates using the uniform and exponential distributions, then convert the point to Cartesian coordinates and the resulting X and Y values will be equivalent to samples generated from N(0, 1).
MATLAB code
In MatLab this algorithm can be implemented with the following code, which generates 20,000 samples from N(0, 1):
x = zeros(10000, 1);
y = zeros(10000, 1);
for ii = 1:10000
u1 = rand;
u2 = rand;
R2 = -2 * log(u1);
theta = 2 * pi * u2;
x(ii) = sqrt(R2) * cos(theta);
y(ii) = sqrt(R2) * sin(theta);
end
hist(x)
In one execution of this script, the following histogram for x was generated:
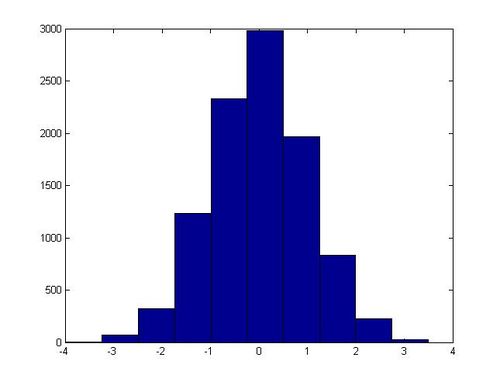
Non-Standard Normal Distributions
Example 1: Single-variate Normal
If X ~ Norm(0, 1) then (a + bX) has a normal distribution with a mean of [math]\displaystyle{ \displaystyle a }[/math] and a standard deviation of [math]\displaystyle{ \displaystyle b }[/math] (which is equivalent to a variance of [math]\displaystyle{ \displaystyle b^2 }[/math]). Using this information with the Box-Muller transform, we can generate values sampled from some random variable [math]\displaystyle{ \displaystyle Y\sim N(a,b^2) }[/math] for arbitrary values of [math]\displaystyle{ \displaystyle a,b }[/math].
- Generate a sample u from Norm(0, 1) using the Box-Muller transform.
- Set v = a + bu.
The values for v generated in this way will be equivalent to sample from a [math]\displaystyle{ \displaystyle N(a, b^2) }[/math]distribution. We can modify the MatLab code used in the last section to demonstrate this. We just need to add one line before we generate the histogram:
v = a + b * x;
For instance, this is the histogram generated when b = 15, a = 125:
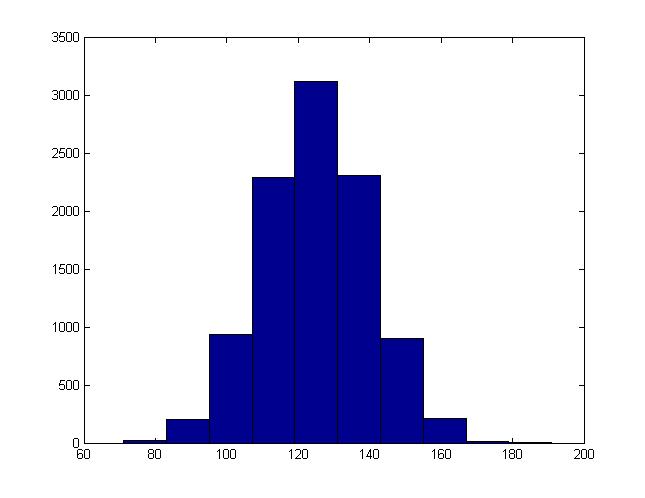
Example 2: Multi-variate Normal
The Box-Muller method can be extended to higher dimensions to generate multivariate normals. The objects generated will be nx1 vectors, and their variance will be described by nxn covariance matrices.
[math]\displaystyle{ \mathbf{z} = N(\mathbf{u}, \Sigma) }[/math] defines the n by 1 vector [math]\displaystyle{ \mathbf{z} }[/math] such that:
- [math]\displaystyle{ \displaystyle u_i }[/math] is the average of [math]\displaystyle{ \displaystyle z_i }[/math]
- [math]\displaystyle{ \!\Sigma_{ii} }[/math] is the variance of [math]\displaystyle{ \displaystyle z_i }[/math]
- [math]\displaystyle{ \!\Sigma_{ij} }[/math] is the co-variance of [math]\displaystyle{ \displaystyle z_i }[/math] and [math]\displaystyle{ \displaystyle z_j }[/math]
If [math]\displaystyle{ \displaystyle z_1, z_2, ..., z_d }[/math] are normal variables with mean 0 and variance 1, then the vector [math]\displaystyle{ \displaystyle (z_1, z_2,..., z_d) }[/math] has mean 0 and variance [math]\displaystyle{ \!I }[/math], where 0 is the zero vector and [math]\displaystyle{ \!I }[/math] is the identity matrix. This fact suggests that the method for generating a multivariate normal is to generate each component individually as single normal variables.
The mean and the covariance matrix of a multivariate normal distribution can be adjusted in ways analogous to the single variable case. If [math]\displaystyle{ \mathbf{z} \sim N(0,I) }[/math], then [math]\displaystyle{ \Sigma^{1/2}\mathbf{z}+\mu \sim N(\mu,\Sigma) }[/math]. Note here that the covariance matrix is symmetric and nonnegative, so its square root should always exist.
We can compute [math]\displaystyle{ \mathbf{z} }[/math] in the following way:
- Generate an n by 1 vector [math]\displaystyle{ \mathbf{x} = \begin{bmatrix}x_{1} & x_{2} & ... & x_{n}\end{bmatrix} }[/math] where [math]\displaystyle{ x_{i} }[/math] ~ Norm(0, 1) using the Box-Muller transform.
- Calculate [math]\displaystyle{ \!\Sigma^{1/2} }[/math] using singular value decomposition.
- Set [math]\displaystyle{ \mathbf{z} = \Sigma^{1/2} \mathbf{x} + \mathbf{u} }[/math].
The following MatLab code provides an example, where a scatter plot of 10000 random points is generated. In this case x and y have a co-variance of 0.9 - a very strong positive correlation.
x = zeros(10000, 1);
y = zeros(10000, 1);
for ii = 1:10000
u1 = rand;
u2 = rand;
R2 = -2 * log(u1);
theta = 2 * pi * u2;
x(ii) = sqrt(R2) * cos(theta);
y(ii) = sqrt(R2) * sin(theta);
end
E = [1, 0.9; 0.9, 1];
[u s v] = svd(E);
root_E = u * (s ^ (1 / 2));
z = (root_E * [x y]');
z(1,:) = z(1,:) + 0;
z(2,:) = z(2,:) + -3;
scatter(z(1,:), z(2,:))
This code generated the following scatter plot:
In Matlab, we can also use the function "sqrtm()" or "chol()" (Cholesky Decomposition) to calculate square root of a matrix directly. Note that the resulting root matrices may be different but this does materially affect the simulation. Here is an example:
E = [1, 0.9; 0.9, 1]; r1 = sqrtm(E); r2 = chol(E);
R code for a multivariate normal distribution:
n=10000; r2<--2*log(runif(n)); theta<-2*pi*(runif(n)); x<-sqrt(r2)*cos(theta); y<-sqrt(r2)*sin(theta); a<-matrix(c(x,y),nrow=n,byrow=F); e<-matrix(c(1,.9,09,1),nrow=2,byrow=T); svde<-svd(e); root_e<-svde$u %*% diag(svde$d)^1/2; z<-t(root_e %*%t(a)); z[,1]=z[,1]+5; z[,2]=z[,2]+ -8; par(pch=19); plot(z,col=rgb(1,0,0,alpha=0.06))
Remarks
MATLAB's randn function uses the ziggurat method to generate normal distributed samples. It is an efficient rejection method based on covering the probability density function with a set of horizontal rectangles so as to obtain points within each rectangle. It is reported that a 800 MHz Pentium III laptop can generate over 10 million random numbers from normal distribution in less than one second. (Reference)
Sampling From Binomial Distributions
In order to generate a sample x from [math]\displaystyle{ \displaystyle X \sim Bin(n, p) }[/math], we can follow the following procedure:
1. Generate n uniform random numbers sampled from [math]\displaystyle{ \displaystyle Unif [0, 1] }[/math]: [math]\displaystyle{ \displaystyle u_1, u_2, ..., u_n }[/math].
2. Set x to be the total number of cases where [math]\displaystyle{ \displaystyle u_i \lt = p }[/math] for all [math]\displaystyle{ \displaystyle 1 \lt = i \lt = n }[/math].
In MatLab this can be coded with a single line. The following generates a sample from [math]\displaystyle{ \displaystyle X \sim Bin(n, p) }[/math]
sum(rand(n, 1) <= p, 1)
Bayesian Inference and Frequentist Inference - October 4, 2011
Bayesian inference vs Frequentist inference
The Bayesian method has become popular in the last few decades as simulation and computer technology makes it more applicable. For more information about its history and application, please refer to http://en.wikipedia.org/wiki/Bayesian_inference. As for frequentist inference, please refer to http://en.wikipedia.org/wiki/Frequentist_inference.
Example
Consider the 'probability' that a person drinks a cup of coffee on a specific day. The interpretations of this for a frequentist and a bayesian are as follows:
Frequentist: There is no explanation to this expression. It is essentially meaningless since it has only occurred once. Therefore, it is not a probability.
Bayesian: Probability captures not only the frequency of occurrences but also one's degree of belief about the random component of a proposition. Therefore it is a valid probability.
Example of face identification
Consider a picture of a face that is associated with an identity (person). Take the face as input x and the person as output y. The person can be either Ali or Tom. We have y=1 if it is Ali and y=0 if it is Tom. We can divide the picture into 100*100 pixels and insert them into a 10,000*1 column vector, which captures x.
If you are a frequentist, you would compare [math]\displaystyle{ \Pr(X=x|y=1) }[/math] with [math]\displaystyle{ \Pr(X=x|y=0) }[/math] and see which one is higher.
If you are Bayesian, you would compare [math]\displaystyle{ \Pr(y=1|X=x) }[/math] with [math]\displaystyle{ \Pr(y=0|X=x) }[/math].
Summary of differences between two schools
- Frequentist: Probability refers to limiting relative frequency. (objective)
- Bayesian: Probability describes degree of belief not frequency. (subjective)
e.g. The probability that you drank a cup of tea on May 20, 2001 is 0.62 does not refer to any frequency.
- Frequentist: Parameters are fixed, unknown constants.
- Bayesian: Parameters are random variables and we can make probabilistic statement about them.
- Frequentist: Statistical procedures should be designed to have long run frequency properties.
e.g. a 95% confidence interval should trap true value of the parameter with limiting frequency at least 95%.
- Bayesian: It makes inferences about [math]\displaystyle{ \theta }[/math] by producing a probability distribution for [math]\displaystyle{ \theta }[/math]. Inference (e.g. point estimates and interval estimates) will be extracted from this distribution :[math]\displaystyle{ f(\theta|X) = \frac{f(X | \theta)\, f(\theta)}{f(X)}. }[/math]
Bayesian inference
Bayesian inference is usually carried out in the following way:
1. Choose a prior probability density function of [math]\displaystyle{ \!\theta }[/math] which is [math]\displaystyle{ f(\!\theta) }[/math]. This is our belief about [math]\displaystyle{ \theta }[/math] before we see any data.
2. Choose a statistical model [math]\displaystyle{ \displaystyle f(x|\theta) }[/math] that reflects our beliefs about X.
3. After observing data [math]\displaystyle{ \displaystyle x_1,...,x_n }[/math], we update our beliefs and calculate the posterior probability.
[math]\displaystyle{ f(\theta|x) = \frac{f(\theta,x)}{f(x)}=\frac{f(x|\theta) \cdot f(\theta)}{f(x)}=\frac{f(x|\theta) \cdot f(\theta)}{\int^{}_\theta f(x|\theta) \cdot f(\theta) d\theta} }[/math], where [math]\displaystyle{ \displaystyle f(\theta|x) }[/math] is the posterior probability, [math]\displaystyle{ \displaystyle f(\theta) }[/math] is the prior probability, [math]\displaystyle{ \displaystyle f(x|\theta) }[/math] is the likelihood of observing X=x given [math]\displaystyle{ \!\theta }[/math] and f(x) is the marginal probability of X=x.
If we have i.i.d. observations [math]\displaystyle{ \displaystyle x_1,...,x_n }[/math], we can replace [math]\displaystyle{ \displaystyle f(x|\theta) }[/math] with [math]\displaystyle{ f({x_1,...,x_n}|\theta)=\prod_{i=1}^n f(x_i|\theta) }[/math] because of independency.
We denote [math]\displaystyle{ \displaystyle f({x_1,...,x_n}|\theta) }[/math] as [math]\displaystyle{ \displaystyle L_n(\theta) }[/math] which is called likelihood. And we use [math]\displaystyle{ \displaystyle x^n }[/math] to denote [math]\displaystyle{ \displaystyle (x_1,...,x_n) }[/math].
[math]\displaystyle{ f(\theta|x^n) = \frac{f(x^n|\theta) \cdot f(\theta)}{f(x^n)}=\frac{f(x^n|\theta) \cdot f(\theta)}{\int^{}_\theta f(x^n|\theta) \cdot f(\theta) d\theta} }[/math] , where [math]\displaystyle{ \int^{}_\theta f(x^n|\theta) \cdot f(\theta) d\theta }[/math] is a constant [math]\displaystyle{ \displaystyle c_n }[/math] which does not depend on [math]\displaystyle{ \displaystyle \theta }[/math]. So [math]\displaystyle{ f(\theta|x^n) \propto f(x^n|\theta) \cdot f(\theta) }[/math]. The posterior probability is proportional to the likelihood times prior probability. Note that it does not matter if we throw away [math]\displaystyle{ \displaystyle c_n }[/math],we can always recover it.
What do we do about the posterier distribution?
- Point estimate
[math]\displaystyle{ \bar{\theta}=\int\theta \cdot f(\theta|x^n) d\theta=\frac{\int\theta \cdot L_n(\theta)\cdot f(\theta) d(\theta)}{c_n} }[/math]
- Baysian Interval estimate
[math]\displaystyle{ \int^{a}_{-\infty} f(\theta|x^n) d\theta=\int^{\infty}_{b} f(\theta|x^n) d\theta=1-\alpha }[/math]
Let C=(a,b); Then [math]\displaystyle{ P(\theta\in C|x^n)=\int^{b}_{a} f(\theta|x^n)d(\theta)=1-\alpha }[/math]. C is a [math]\displaystyle{ \displaystyle 1-\alpha }[/math] posterior interval.
Let [math]\displaystyle{ \tilde{\theta}=(\theta_1,...,\theta_d)^T }[/math], then [math]\displaystyle{ f(\theta_1|x^n) = \int^{} \int^{} \dots \int^{}f(\tilde{\theta}|X)d\theta_2d\theta_3 \dots d\theta_d }[/math] and [math]\displaystyle{ E(\theta_1|x^n)=\int^{}\theta_1 \cdot f(\theta_1|x^n) d\theta_1 }[/math]
Example 1: Estimating parameters of a univariate Gaussian distribution
Suppose X follows a univariate Gaussian distribution (i.e. a Normal distribution) with parameters [math]\displaystyle{ \!\mu }[/math] and [math]\displaystyle{ \displaystyle {\sigma^2} }[/math].
(a) For Frequentists:
[math]\displaystyle{ f(x|\theta)= \frac{1}{\sqrt{2\pi}\sigma} \cdot e^{-\frac{1}{2}(\frac{x-\mu}{\sigma})^2} }[/math]
[math]\displaystyle{ L_n(\theta)= \prod_{i=1}^n \frac{1}{\sqrt{2\pi}\sigma} \cdot e^{-\frac{1}{2}(\frac{x_i-\mu}{\sigma})^2} }[/math]
[math]\displaystyle{ \ln L_n(\theta) = l(\theta) = \sum_{i=1}^n[ -\frac{1}{2}\ln 2\pi-\ln \sigma-\frac{1}{2}(\frac{x_i-\mu}{\sigma})^2] }[/math]
To get the maximum likelihood estimator of [math]\displaystyle{ \!\mu }[/math] (mle), we find the [math]\displaystyle{ \hat{\mu} }[/math] which maximizes [math]\displaystyle{ \displaystyle L_n(\theta) }[/math]:
[math]\displaystyle{ \frac{\partial l(\theta)}{\partial \mu}= \sum_{i=1}^n \frac{1}{\sigma}(\frac{x_i-\mu}{\sigma})=0 \Rightarrow \sum_{i=1}^n x_i = n\mu \Rightarrow \hat{\mu}_{mle}=\bar{x} }[/math]
(b) For Bayesians:
[math]\displaystyle{ f(\theta|x) \propto f(x|\theta) \cdot f(\theta) }[/math]
We assume that the mean of the above normal distribution is itself distributed normally with mean [math]\displaystyle{ \!\mu_0 }[/math] and variance [math]\displaystyle{ \!\Gamma }[/math].
Suppose [math]\displaystyle{ \!\mu\sim N(\mu_0, \!\Gamma^2 }[/math]),
so [math]\displaystyle{ f(\mu) = \frac{1}{\sqrt{2\pi}\Gamma} \cdot e^{-\frac{1}{2}(\frac{\mu-\mu_0}{\Gamma})^2} }[/math]
[math]\displaystyle{ f(x|\mu) = \frac{1}{\sqrt{2\pi}\tilde{\sigma}} \cdot e^{-\frac{1}{2}(\frac{x-\mu}{\tilde{\sigma}})^2} }[/math]
[math]\displaystyle{ \tilde{\mu} = \frac{\frac{n}{\sigma^2}}{\frac{n}{\sigma^2}+\frac{1}{\Gamma^2}}\bar{x}+\frac{\frac{1}{\Gamma^2}}{\frac{n}{\sigma^2}+\frac{1}{\Gamma^2}}\mu_0 }[/math], where [math]\displaystyle{ \tilde{\mu} }[/math] is the estimator of [math]\displaystyle{ \!\mu }[/math].
- If prior belief about [math]\displaystyle{ \!\mu_0 }[/math] is strong, then [math]\displaystyle{ \!\Gamma }[/math] is small and [math]\displaystyle{ \frac{1}{\Gamma^2} }[/math] is large. [math]\displaystyle{ \tilde{\mu} }[/math] is close to [math]\displaystyle{ \!\mu_0 }[/math] and the observations will not affect too much. On the contrary, if prior belief about [math]\displaystyle{ \!\mu_0 }[/math] is weak, [math]\displaystyle{ \!\Gamma }[/math] is large and [math]\displaystyle{ \frac{1}{\Gamma^2} }[/math] is small. [math]\displaystyle{ \tilde{\mu} }[/math] depends more on observations. (This is intuitive, when our original belief is reliable, then the sample is not important in improving the result; when the belief is not reliable, then we depend a lot on the sample.)
- When the sample is large (i.e. n [math]\displaystyle{ \to \infty }[/math]), [math]\displaystyle{ \tilde{\mu} \to \bar{x} }[/math] and the impact of prior belief about [math]\displaystyle{ \!\mu }[/math] is weakened.
Basic Monte Carlo Integration - October 6th, 2011
Three integration methods would be taught in this course:
- Basic Monte Carlo Integration
- Importance Sampling
- Markov Chain Monte Carlo (MCMC)
The first, and most basic, method of numerical integration we will see is Monte Carlo Integration. We use this to solve an integral of the form: [math]\displaystyle{ I = \int_{a}^{b} h(x) dx }[/math]
Note the following derivation:
[math]\displaystyle{ \begin{align} \displaystyle I & = \int_{a}^{b} h(x)dx \\ & = \int_{a}^{b} h(x)((b-a)/(b-a))dx \\ & = \int_{a}^{b} (h(x)(b-a))(1/(b-a))dx \\ & = \int_{a}^{b} w(x)f(x)dx \\ & = E[w(x)] \\ \end{align} }[/math]
~ [math]\displaystyle{ \frac{1}{n} \sum_{i=1}^{n} w(x) }[/math]
Where w(x) = h(x)(b-a) and f(x) is the probability density function of a uniform random variable on the interval [a,b]. The expectation, with respect to the distribution of f, of w is taken from n samples of x.
General Procedure
i) Draw n samples [math]\displaystyle{ x_i \sim~ Unif[a,b] }[/math]
ii) Compute [math]\displaystyle{ \ w(x_i) }[/math] for every sample
iii) Obtain an estimate of the integral, [math]\displaystyle{ \hat{I} }[/math], as follows:
[math]\displaystyle{ \hat{I} = \frac{1}{n} \sum_{i=1}^{n} w(x_i) }[/math] . Clearly, this is just the average of the simulation results.
By the strong law of large numbers [math]\displaystyle{ \hat{I} }[/math] converges to [math]\displaystyle{ \ I }[/math] as [math]\displaystyle{ \ n \rightarrow \infty }[/math]. Because of this, we can compute all sorts of useful information, such as variance, standard error, and confidence intervals.
Standard Error: [math]\displaystyle{ SE = \frac{Standard Deviation} {\sqrt{n}} }[/math]
Variance: [math]\displaystyle{ V = \frac{\sum_{i=1}^{n} (w(x)-\hat{I})^2}{n-1} }[/math]
Confidence Interval: [math]\displaystyle{ \hat{I} \pm t_{(\alpha/2)} SE }[/math]
Example: Uniform Distribution
Consider the integral, [math]\displaystyle{ \int_{0}^{1} x^3dx }[/math], which is easily solved through standard analytical integration methods, and is equal to .25. Now, let us check this answer with a numerical approximation using Monte Carlo Integration.
We generate a 1 by 10000 vector of uniform (on the interval [0,1]) random variables and call that vector 'u'. We see that our 'w' in this case is [math]\displaystyle{ x^3 }[/math], so we set [math]\displaystyle{ w = u^3 }[/math]. Our [math]\displaystyle{ \hat{I} }[/math] is equal to the mean of w.
In Matlab, we can solve this integration problem with the following code:
u = rand(1,10000); w = u.^3; mean(w) ans = 0.2475
Note the '.' after 'u' in the second line of code, indicating that each entry in the matrix is cubed. Also, our approximation is close to the actual value of .25. Now let's try to get an even better approximation by generating more sample points.
u= rand(1,100000); w= u.^3; mean(w) ans = .2503
We see that when the number of sample points is increased, our approximation improves, as one would expect.
Generalization
Up to this point we have seen how to numerically approximate an integral when the distribution of f is uniform. Now we will see how to generalize this to other distributions.
[math]\displaystyle{ I = \int h(x)f(x)dx }[/math]
If f is a distribution function (pdf), then [math]\displaystyle{ I }[/math] can be estimated as Ef[h(x)]. This means taking the expectation of h with respect to the distribution of f. Our previous example is the case where f is the uniform distribution between [a,b].
Procedure for the General Case
i) Draw n samples from f
ii) Compute h(xi)
iii) [math]\displaystyle{ \hat{I} = \frac{1}{n} \sum_{i=1}^{n} h(x }[/math]i[math]\displaystyle{ ) }[/math]
Example: Exponential Distribution
Find [math]\displaystyle{ E[\sqrt{x}] }[/math] for [math]\displaystyle{ \displaystyle f = e^{-x} }[/math], which is the exponential distribution with mean 1.
[math]\displaystyle{ I = \int_{0}^{\infty} \sqrt{x} e^{-x}dx }[/math]
We can see that we must draw samples from f, the exponential distribution.
To find a numerical solution using Monte Carlo Integration we see that:
u= rand(1,10000)
X= -log(u)
h= [math]\displaystyle{ \sqrt{x} }[/math]
I= mean(h)
To implement this procedure in Matlab, use the following code:
u = rand(1,10000); X = -log(u); h = x.^.5; mean(h) ans = .8841
An easy way to check whether your approximation is correct is to use the built in Matlab function 'quadl' which takes a function and bounds for the integral and returns a solution for the definite integral of that function. For this specific example, we can enter:
f = @(x) sqrt(x).*exp(-x); % quadl runs into computational problems when the upper bound is "inf" or an extremely large number, % so choose just a moderately large number. quadl(f,0,100) ans = 0.8862
From the above result, we see that our approximation was quite close.
Example: Normal Distribution
Let [math]\displaystyle{ f(x) = (1/(2 \pi)^{1/2}) e^{(-x^2)/2} }[/math]. Compute the cumulative distribution function at some point x.
[math]\displaystyle{ F(x)= \int_{-\infty}^{x} f(s)ds = \int_{-\infty}^{x}(1)(1/(2 \pi)^{1/2}) e^{(-s^2)/2}ds }[/math]. The (1) is inserted to illustrate that our h(x) will be the constant function 1, and our f(x) is the normal distribution. To take into account the upper bound of integration, x, any values sampled that are greater than x will be set to zero.
This is the Matlab code for solving F(2):
u = randn(1,10000) h = u < 2; mean(h) ans = .9756
We generate a 1 by 10000 vector of standard normal random variables and we return a value of 1 if u is less than 2, and 0 otherwise.
We can also build the function F(x) in matlab in the following way:
function F(x) u=rand(1,1000000); h=u<x; mean(h)
Example: Binomial Distribution
In this example we will see the Bayesian Inference for 2 Binomial Distributions.
Let [math]\displaystyle{ X ~ Bin(n,p) }[/math] and [math]\displaystyle{ Y ~ Bin(m,q) }[/math], and let [math]\displaystyle{ \!\delta = p-q }[/math].
Therefore, [math]\displaystyle{ \displaystyle \!\delta = x/n - y/m }[/math] which is the frequentist approach.
Bayesian wants [math]\displaystyle{ \displaystyle f(p,q|x,y) = f(x,y|p,q)f(p,q)/f(x,y) }[/math], where [math]\displaystyle{ f(x,y)=\iint\limits_{\!\theta} f(x,y|p,q)f(p,q)\,dp\,dq }[/math] is a constant.
Thus, [math]\displaystyle{ \displaystyle f(p,q|x,y)\propto f(x,y|p,q)f(p,q) }[/math]. Now we assume that [math]\displaystyle{ \displaystyle f(p,q) = f(p)f(q) = 1 }[/math] and f(p) and f(q) are uniform.
Therefore, [math]\displaystyle{ \displaystyle f(p,q|x,y)\propto p^x(1-p)^{n-x}q^y(1-q)^{m-y} }[/math].
[math]\displaystyle{ E[\delta|x,y] = \int_{0}^{1} \int_{0}^{1} (p-q)f(p,q|x,y)dpdq }[/math].
As you can see this is much tougher than the frequentist approach.
Importance Sampling and Basic Monte Carlo Integration - October 11th, 2011
Example: Binomial Distribution (Continued)
Suppose we are given two independent Binomial Distributions [math]\displaystyle{ \displaystyle X \sim Bin(n, p_1) }[/math], [math]\displaystyle{ \displaystyle Y \sim Bin(m, p_2) }[/math]. We would like to give an Monte Carlo estimate of [math]\displaystyle{ \displaystyle \delta = p_1 - p_2 }[/math]
Frequentist approach:
[math]\displaystyle{ \displaystyle \hat{p_1} = \frac{X}{n} }[/math] ; [math]\displaystyle{ \displaystyle \hat{p_2} = \frac{Y}{m} }[/math]
[math]\displaystyle{ \displaystyle \hat{\delta} = \hat{p_1} - \hat{p_2} = \frac{X}{n} - \frac{Y}{m} }[/math]
Bayesian approach to compute the expected value of [math]\displaystyle{ \displaystyle \delta }[/math]:
[math]\displaystyle{ \displaystyle E(\delta|X,Y) = \int\int(p_1-p_2) f(p_1,p_2|X,Y)\,dp_1dp_2 }[/math]
Assume that [math]\displaystyle{ \displaystyle n = 100, m = 100, p_1 = 0.5, p_2 = 0.8 }[/math] and the sample size is 1000.
MATLAB code of the above example:
n = 100; m = 100; p_1 = 0.5; p_2 = 0.8; p1 = mean(rand(n,1000)<p_1,1); p2 = mean(rand(m,1000)<p_2,1); delta = p2 - p1; hist(delta) mean(delta)
In one execution of the code, the mean of delta was 0.3017. The histogram of delta generated was:
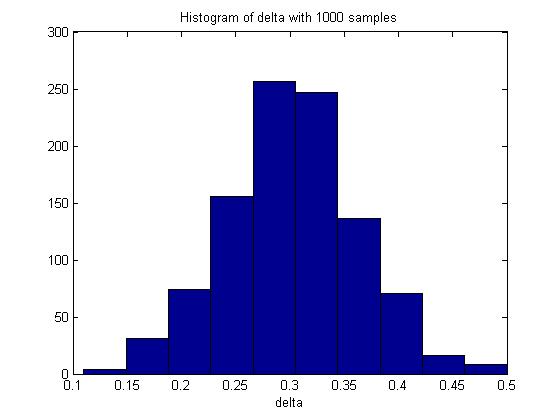
Through Monte Carlo simulation, we can obtain an empirical distribution of delta and carry out inference on the data obtained, such as computing the mean, maximum, variance, standard deviation and the standard error of delta.
Importance Sampling
Motivation
Consider the integral [math]\displaystyle{ \displaystyle I = \int h(x)f(x)\,dx }[/math]
According to basic Monte Carlo Integration, if we can sample from the probability density function [math]\displaystyle{ \displaystyle f(x) }[/math] and feed the samples of [math]\displaystyle{ \displaystyle f(x) }[/math] back to [math]\displaystyle{ \displaystyle h(x) }[/math], [math]\displaystyle{ \displaystyle I }[/math] can be estimated as an average of [math]\displaystyle{ \displaystyle h(x) }[/math] ( i.e. [math]\displaystyle{ \hat{I} = \frac{1}{n} \sum_{i=1}^{n} h(x_i) }[/math] )
However, the Monte Carlo method works when we know how to sample from [math]\displaystyle{ \displaystyle f(x) }[/math]. In the case where it is difficult to sample from [math]\displaystyle{ \displaystyle f(x) }[/math], importance sampling is a technique that we can apply. Importance Sampling relies on another function [math]\displaystyle{ \displaystyle g(x) }[/math] which we know how to sample from.
The above integral can be rewritten as follow:
[math]\displaystyle{ \begin{align}
\displaystyle I & = \int h(x)f(x)\,dx \\
& = \int h(x)f(x)\frac{g(x)}{g(x)}\,dx \\
& = \int \frac{h(x)f(x)}{g(x)}g(x)\,dx \\
& = \int y(x)g(x)\,dx \\
& = E_g(y(x)) \\
\end{align}
}[/math]
[math]\displaystyle{ where \ y(x) = \frac{h(x)f(x)}{g(x)} }[/math]
The integral can thus be simulated as [math]\displaystyle{ \displaystyle \hat{I} = \frac{1}{n} \sum_{i=1}^{n} Y_i \ , \ where \ Y_i = \frac{h(x_i)f(x_i)}{g(x_i)} }[/math]
Procedure
Suppose we know how to sample from [math]\displaystyle{ \displaystyle g(x) }[/math]
- Choose a suitable [math]\displaystyle{ \displaystyle g(x) }[/math] and draw n samples [math]\displaystyle{ x_1,x_2....,x_n \sim~ g(x) }[/math]
- Set [math]\displaystyle{ Y_i =\frac{h(x_i)f(x_i)}{g(x_i)} }[/math]
- Compute [math]\displaystyle{ \hat{I} = \frac{1}{n}\sum_{i=1}^{n} Y_i }[/math]
By the Law of large numbers, [math]\displaystyle{ \displaystyle \hat{I} \rightarrow I }[/math] provided that the sample size n is large enough.
Remarks: One can think of [math]\displaystyle{ \frac{f(x)}{g(x)} }[/math] as a weight to [math]\displaystyle{ \displaystyle h(x) }[/math] in the computation of [math]\displaystyle{ \hat{I} }[/math]
[math]\displaystyle{ \displaystyle i.e. \ \hat{I} = \frac{1}{n}\sum_{i=1}^{n} Y_i = \frac{1}{n}\sum_{i=1}^{n} (\frac{f(x_i)}{g(x_i)})h(x_i) }[/math]
Therefore, [math]\displaystyle{ \displaystyle \hat{I} }[/math] is a weighted average of [math]\displaystyle{ \displaystyle h(x_i) }[/math]
Problem
If [math]\displaystyle{ \displaystyle g(x) }[/math] is not chosen appropriately, then the variance of the estimate [math]\displaystyle{ \hat{I} }[/math] may be very large. The problem here is actually similar to what we encounter with the Rejection-Acceptance method. Consider the second moment of [math]\displaystyle{ y(x) }[/math]:
[math]\displaystyle{ \begin{align}
E_g((y(x))^2) \\
& = \int (\frac{h(x)f(x)}{g(x)})^2 g(x) dx \\
& = \int \frac{h^2(x) f^2(x)}{g^2(x)} g(x) dx \\
& = \int \frac{h^2(x)f^2(x)}{g(x)} dx \\
\end{align}
}[/math]
When [math]\displaystyle{ \displaystyle g(x) }[/math] is very small, then the above integral could be very large, hence the variance can be very large when g is not chosen appropriately. This occurs when [math]\displaystyle{ \displaystyle g(x) }[/math] has a thinner tail than [math]\displaystyle{ \displaystyle f(x) }[/math] such that the quantity [math]\displaystyle{ \displaystyle \frac{h^2(x)f^2(x)}{g(x)} }[/math] is large.
Remarks:
1. We can actually compute the form of [math]\displaystyle{ \displaystyle g(x) }[/math] to have optimal variance.
Mathematically, it is to find [math]\displaystyle{ \displaystyle g(x) }[/math] subject to [math]\displaystyle{ \displaystyle \min_g [\ E_g([y(x)]^2) - (E_g[y(x)])^2\ ] }[/math]
It can be shown that the optimal [math]\displaystyle{ \displaystyle g(x) }[/math] is [math]\displaystyle{ \displaystyle {|h(x)|f(x)} }[/math]. Using the optimal [math]\displaystyle{ \displaystyle g(x) }[/math] will minimize the variance of estimation in Importance Sampling. This is of theoretical interest but not useful in practice. As we can see, if we can actually show the expression of g(x), we must first have the value of the integration---which is what we want in the first place.
2. In practice, we shall choose [math]\displaystyle{ \displaystyle g(x) }[/math] which has similar shape as [math]\displaystyle{ \displaystyle f(x) }[/math] but with a thicker tail than [math]\displaystyle{ \displaystyle f(x) }[/math] in order to avoid the problem mentioned above.
Example
Estimate [math]\displaystyle{ \displaystyle I = Pr(Z\gt 3),\ where\ Z \sim N(0,1) }[/math]
Method 1: Basic Monte Carlo
[math]\displaystyle{ \begin{align} Pr(Z\gt 3) & = \int^\infty_3 f(x)\,dx \\
& = \int^\infty_{-\infty} h(x)f(x)\,dx \end{align} }[/math]
[math]\displaystyle{ where \
h(x) = \begin{cases}
0, & \text{if } x \le 3 \\
1, & \text{if } x \gt 3
\end{cases} }[/math]
[math]\displaystyle{ \ ,\ f(x) = \frac{1}{\sqrt{2\pi}}e^{-\frac{1}{2}x^2} }[/math]
MATLAB code to compute [math]\displaystyle{ \displaystyle I }[/math] from 100 samples of standard normal distribution:
h = randn(100,1) > 3; I = mean(h)
In one execution of the code, it returns a value of 0 for [math]\displaystyle{ \displaystyle I }[/math], which differs significantly from the true value of [math]\displaystyle{ \displaystyle I \approx 0.0013 }[/math]. The problem of using Basic Monte Carlo in this example is that [math]\displaystyle{ \displaystyle Pr(Z\gt 3) }[/math] has a small value, and hence many points sampled from the standard normal distribution will be wasted. Therefore, although Basic Monte Carlo is a feasible method to compute [math]\displaystyle{ \displaystyle I }[/math], it gives a poor estimation.
Method 2: Importance Sampling
[math]\displaystyle{ \displaystyle I = Pr(Z\gt 3)= \int^\infty_3 f(x)\,dx }[/math]
To apply importance sampling, we have to choose a [math]\displaystyle{ \displaystyle g(x) }[/math] which we will sample from. In this example, we can choose [math]\displaystyle{ \displaystyle g(x) }[/math] to be the probability density function of exponential distribution, normal distribution with mean 0 and variance greater than 1 or normal distribution with mean greater than 0 and variance 1 etc. The goal is to minimize the number of rejected samples in order to produce a more accurate result! For the following, we take [math]\displaystyle{ \displaystyle g(x) }[/math] to be the pdf of [math]\displaystyle{ \displaystyle N(4,1) }[/math].
Procedure:
- Draw n samples [math]\displaystyle{ x_1,x_2....,x_n \sim~ g(x) }[/math]
- Calculate [math]\displaystyle{ \begin{align} \frac{f(x)}{g(x)} & = \frac{ \frac{1}{\sqrt{2\pi}}e^{-\frac{1}{2}x^2}
}{ \frac{1}{\sqrt{2\pi}}e^{-\frac{1}{2}(x-4)^2} } \\
& = e^{8-4x} \end{align} }[/math]
- Set [math]\displaystyle{ Y_i = h(x_i)e^{8-4x_i}\ with\ h(x) = \begin{cases}
0, & \text{if } x \le 3 \\
1, & \text{if } x \gt 3
\end{cases}
}[/math]
- Compute [math]\displaystyle{ \hat{Y} = \frac{1}{n}\sum_{i=1}^{n} Y_i }[/math]
The above procedure from 100 samples of [math]\displaystyle{ \displaystyle g(x) }[/math]can be implemented in MATLAB as follow:
for ii = 1:100 x = randn + 4 ; h = x > 3 ; y(ii) = h * exp(8-4*x) ; end mean(y)
In one execution of the code, it returns a value of 0.001271 for [math]\displaystyle{ \hat{Y} }[/math], which is much closer to the true value of [math]\displaystyle{ \displaystyle I \approx 0.0013 }[/math]. From many executions of the code, the variance of basic monte carlo is approximately 150 times that of importance sampling. This demonstrates that this method can provide a better estimate than the Basic Monte Carlo method.
Importance Sampling with Normalized Weight and Markov Chain Monte Carlo - October 13th, 2011
Importance Sampling with Normalized Weight
Recall that we can think of [math]\displaystyle{ \displaystyle b(x) = \frac{f(x)}{g(x)} }[/math] as a weight applied to the samples [math]\displaystyle{ \displaystyle h(x) }[/math]. If the form of [math]\displaystyle{ \displaystyle f(x) }[/math] is known only up to a constant, we can use an alternate, normalized form of the weight, [math]\displaystyle{ \displaystyle b^*(x) }[/math]. (This situation arises in Bayesian inference.) Importance sampling with normalized or standard weight is also called indirect importance sampling.
We derive the normalized weight as follows:
[math]\displaystyle{ \begin{align}
\displaystyle I & = \int h(x)f(x)\,dx \\
&= \int h(x)\frac{f(x)}{g(x)}g(x)\,dx \\
&= \frac{\int h(x)\frac{f(x)}{g(x)}g(x)\,dx}{\int f(x) dx} \\
&= \frac{\int h(x)\frac{f(x)}{g(x)}g(x)\,dx}{\int \frac{f(x)}{g(x)}g(x) dx} \\
&= \frac{\int h(x)b(x)g(x)\,dx}{\int\ b(x)g(x) dx}
\end{align} }[/math]
[math]\displaystyle{ \hat{I}= \frac{\sum_{i=1}^{n} h(x_i)b(x_i)}{\sum_{i=1}^{n} b(x_i)} }[/math]
Then, the normalized weight is [math]\displaystyle{ b^*(x) = \displaystyle \frac{b(x_i)}{\sum_{i=1}^{n} b(x_i)} }[/math]
Note that [math]\displaystyle{ \int f(x) dx = 1 = \int b(x)g(x) dx = 1 }[/math]
We can also determine the associated Monte Carlo variance of this estimate by
[math]\displaystyle{ Var(\hat{I})= \frac{\sum_{i=1}^{n} b(x_i)(h(x_i) - \hat{I})^2}{\sum_{i=1}^{n} b(x_i)} }[/math]
Markov Chain Monte Carlo
We still want to solve [math]\displaystyle{ I = \displaystyle\int^\ h(x)f(x)\,dx }[/math]
Stochastic Process
A stochastic process [math]\displaystyle{ \{ x_t : t \in T \} }[/math] is a collection of random variables. Variables [math]\displaystyle{ \displaystyle x_t }[/math] take values in some set [math]\displaystyle{ \displaystyle X }[/math] called the state space. The set [math]\displaystyle{ \displaystyle T }[/math] is called the index set.
Markov Chain
A Markov Chain is a stochastic process for which the distribution of [math]\displaystyle{ \displaystyle x_t }[/math] depends only on [math]\displaystyle{ \displaystyle x_{t-1} }[/math]. It is a random process characterized as being memoryless, meaning that the next occurrence of a defined event is only dependent on the current event;not on the preceding sequence of events.
Formal Definition: The process [math]\displaystyle{ \{ x_t : t \in T \} }[/math] is a Markov Chain if [math]\displaystyle{ \displaystyle Pr(x_t|x_0, x_1,..., x_{t-1})= Pr(x_t|x_{t-1}) }[/math] for all [math]\displaystyle{ \{t \in T \} }[/math] and for all [math]\displaystyle{ \{x \in X \} }[/math]
For a Markov Chain, [math]\displaystyle{ \displaystyle f(x_1,...x_n)= f(x_1)f(x_2|x_1)f(x_3|x_2)...f(x_n|x_{n-1}) }[/math]
Real Life Example:
When going for an interview, the employer only looks at your highest education achieved. The employer would not look at the past educations received (elementary school, high school etc.) because the employer believes that the highest education achieved summarizes your previous educations. Therefore, anything before your most recent previous education is irrelevant. In other word, we can say that [math]\displaystyle{ \! x_t }[/math] is regarded as the summary of [math]\displaystyle{ \!x_{t-1},...,x_2,x_1 }[/math], so when we need to determine [math]\displaystyle{ \!x_{t+1} }[/math], we only need to pay attention in [math]\displaystyle{ \!x_{t} }[/math].
Transition Probabilities
The transition probability is the probability of jumping from one state to another state in a Markov Chain.
Formally, let us define [math]\displaystyle{ \displaystyle P_{ij} = Pr(x_{t+1}=j|x_t=i) }[/math] to be the transition probability.
That is, [math]\displaystyle{ \displaystyle P_{ij} }[/math] is the probability of transitioning from state i to state j in a single step. Then the matrix [math]\displaystyle{ \displaystyle P }[/math] whose (i,j) element is [math]\displaystyle{ \displaystyle P_{ij} }[/math] is called the transition matrix.
Properties of P:
- 1) [math]\displaystyle{ \displaystyle P_{ij} \gt = 0 }[/math] (The probability of going to another state cannot be negative)
- 2) [math]\displaystyle{ \displaystyle \sum_{\forall j}P_{ij} = 1 }[/math] (The probability of transitioning to some state from state i (including remaining in state i) is one)
Random Walk
Example: Start at one point and flip a coin where [math]\displaystyle{ \displaystyle Pr(H)=p }[/math] and [math]\displaystyle{ \displaystyle Pr(T)=1-p=q }[/math]. Take one step right if heads and one step left if tails. If at an endpoint, stay there. The transition matrix is [math]\displaystyle{ P=\left(\begin{matrix}1&0&0&\dots&\dots&0\\ q&0&p&0&\dots&0\\ 0&q&0&p&\dots&0\\ \vdots&\vdots&\vdots&\ddots&\vdots&\vdots\\ \vdots&\vdots&\vdots&\vdots&\ddots&\vdots\\ 0&0&\dots&\dots&\dots&1 \end{matrix}\right) }[/math]
Let [math]\displaystyle{ \displaystyle P_n }[/math] be the matrix such that its (i,j) element is [math]\displaystyle{ \displaystyle P_{ij}(n) }[/math]. This is called n-step probability.
- [math]\displaystyle{ \displaystyle P_n = P^n }[/math]
- [math]\displaystyle{ \displaystyle P_1 = P }[/math]
- [math]\displaystyle{ \displaystyle P_2 = P^2 }[/math]
Markov Chain Properties and Page Rank - October 18th, 2011
Summary of Terminology
Transition Matrix
A matrix [math]\displaystyle{ \!P }[/math] that defines a Markov Chain has the form:
[math]\displaystyle{ P = \begin{bmatrix} P_{11} & \cdots & P_{1N} \\ \vdots & \ddots & \vdots \\ P_{N1} & \cdots & P_{NN} \end{bmatrix} }[/math]
where [math]\displaystyle{ \!P(i,j) = P_{ij} = Pr(x_{t+1} = j | x_t = i) }[/math] is the probability of transitioning from state i to state j in the Markov Chain in a single step. Note that this implies that all rows add up to one.
n-step Transition matrix
A matrix [math]\displaystyle{ \!P_n }[/math] whose (i,j)th entry is the probability of moving from state i to state j after n transitions:
[math]\displaystyle{ \!P_n(i,j) = Pr(x_{m+n}=j|x_m = i) }[/math]
This probability is called the n-step transition probability. A nice property of this matrix is that
[math]\displaystyle{ \!P_n = P^n }[/math]
For all n >= 0, where P is the transition matrix. Note that the rows of [math]\displaystyle{ \!P_n }[/math] should still add up to one.
Marginal distribution of a Markov Chain
We represent the state at time t as a vector.
[math]\displaystyle{ \mu_t = (\mu_t(1) \; \mu_t(2) \; ... \; \mu_t(n)) }[/math]
Consider this Markov Chain:
[math]\displaystyle{ \mu_t = (A \; B) }[/math], where A is the probability of being in state a at time t, and B is the probability of being in state b at time t.
For example if [math]\displaystyle{ \mu_t = (0.1 \; 0.9) }[/math], we have a 10% chance of being in state a at time t, and a 90% chance of being in state b at time t.
Suppose we run this Markov chain many times, and record the state at each step.
In this example, we run 4 trials, up until t=5.
| t | Trial 1 | Trial 2 | Trial 3 | Trial 4 | Observed [math]\displaystyle{ \mu }[/math] |
|---|---|---|---|---|---|
| 1 | a | b | b | a | (0.5, 0.5) |
| 2 | b | a | a | a | (0.75, 0.25) |
| 3 | a | a | b | a | (0.75, 0.25) |
| 4 | b | b | a | b | (0.25, 0.75) |
| 5 | b | b | b | a | (0.25, 0.75) |
Imagine simulating the chain many times. If we collect all the outcomes at time t from all the chains, the histogram of this data would look like [math]\displaystyle{ \!\mu_t }[/math].
We can find the marginal probabilities as [math]\displaystyle{ \!\mu_n = \mu_0 P^n }[/math]
Stationary Distribution
Let [math]\displaystyle{ \pi = (\pi_i \mid i \in \chi) }[/math] be a vector of non-negative numbers that sum to 1. (i.e. [math]\displaystyle{ \!\pi }[/math] is a pmf)
If [math]\displaystyle{ \!\pi = \pi P }[/math], then [math]\displaystyle{ \!\pi }[/math] is a stationary distribution, also known as an invariant distribution.
Limiting Distribution
A Markov chain has limiting distribution [math]\displaystyle{ \!\pi }[/math] if [math]\displaystyle{ \lim_{n \to \infty} P^n = \begin{bmatrix} \pi \\ \vdots \\ \pi \end{bmatrix} }[/math]
That is, [math]\displaystyle{ \!\pi_j = \lim_{n \to \infty}\left [ P^n \right ]_{ij} }[/math] exists and is independent of i.
Here is an example:
Suppose we want to find the stationary distribution of [math]\displaystyle{ P=\left(\begin{matrix} 1/3&1/3&1/3\\ 1/4&3/4&0\\ 1/2&0&1/2 \end{matrix}\right) }[/math]
We want to solve [math]\displaystyle{ \pi=\pi P }[/math] and we want [math]\displaystyle{ \displaystyle \pi_0 + \pi_1 + \pi_2 = 1 }[/math]
[math]\displaystyle{ \displaystyle \pi_0 = \frac{1}{3}\pi_0 + \frac{1}{4}\pi_1 + \frac{1}{2}\pi_2 }[/math]
[math]\displaystyle{ \displaystyle \pi_1 = \frac{1}{3}\pi_0 + \frac{3}{4}\pi_1 }[/math]
[math]\displaystyle{ \displaystyle \pi_2 = \frac{1}{3}\pi_0 + \frac{1}{2}\pi_2 }[/math]
Solving the system of equations, we get
[math]\displaystyle{ \displaystyle \pi_1 = \frac{4}{3}\pi_0 }[/math]
[math]\displaystyle{ \displaystyle \pi_2 = \frac{2}{3}\pi_0 }[/math]
So using our condition above, we have [math]\displaystyle{ \displaystyle \pi_0 + \frac{4}{3}\pi_0 + \frac{2}{3}\pi_0 = 1 }[/math] and by solving we get [math]\displaystyle{ \displaystyle \pi_0 = \frac{1}{3} }[/math]
Using this in our system of equations, we obtain:
[math]\displaystyle{ \displaystyle \pi_1 = \frac{4}{9} }[/math]
[math]\displaystyle{ \displaystyle \pi_2 = \frac{2}{9} }[/math]
Thus, the stationary distribution is [math]\displaystyle{ \displaystyle \pi = (\frac{1}{3}, \frac{4}{9}, \frac{2}{9}) }[/math]
Detailed Balance
[math]\displaystyle{ \!\pi }[/math] has the detailed balance property if [math]\displaystyle{ \!\pi_iP_{ij} = P_{ji}\pi_j }[/math]
Theorem
If [math]\displaystyle{ \!\pi }[/math] satisfies detailed balance, then [math]\displaystyle{ \!\pi }[/math] is a stationary distribution.
In other words, if [math]\displaystyle{ \!\pi_iP_{ij} = P_{ji}\pi_j }[/math], then [math]\displaystyle{ \!\pi = \pi P }[/math]
Proof:
[math]\displaystyle{ \!\pi P = \begin{bmatrix}\pi_1 & \pi_2 & \cdots & \pi_N\end{bmatrix} \begin{bmatrix}P_{11} & \cdots & P_{1N} \\ \vdots & \ddots & \vdots \\ P_{N1} & \cdots & P_{NN}\end{bmatrix} }[/math]
Observe that the jth element of [math]\displaystyle{ \!\pi P }[/math] is
[math]\displaystyle{ \!\left [ \pi P \right ]_j = \pi_1 P_{1j} + \pi_2 P_{2j} + \dots + \pi_N P_{Nj} }[/math]
- [math]\displaystyle{ \! = \sum_{i=1}^N \pi_i P_{ij} }[/math]
- [math]\displaystyle{ \! = \sum_{i=1}^N P_{ji} \pi_j }[/math], by the definition of detailed balance.
- [math]\displaystyle{ \! = \pi_j \sum_{i=1}^N P_{ji} }[/math]
- [math]\displaystyle{ \! = \pi_j }[/math], as the sum of the entries in a row of P must sum to 1.
So [math]\displaystyle{ \!\pi = \pi P }[/math].
Example
Find the marginal distribution of
Start by generating the matrix P.
[math]\displaystyle{ \!P = \begin{pmatrix} 0.2 & 0.8 \\ 0.6 & 0.4 \end{pmatrix} }[/math]
We must assume some starting value for [math]\displaystyle{ \mu_0 }[/math]
[math]\displaystyle{ \!\mu_0 = \begin{pmatrix} 0.1 & 0.9 \end{pmatrix} }[/math]
For t = 1, the marginal distribution is
[math]\displaystyle{ \!\mu_1 = \mu_0 P }[/math]
Notice that this [math]\displaystyle{ \mu }[/math] converges.
If you repeatedly run:
[math]\displaystyle{ \!\mu_{i+1} = \mu_i P }[/math]
It converges to [math]\displaystyle{ \mu = \begin{pmatrix} 0.4286 & 0.5714 \end{pmatrix} }[/math]
This can be seen by running the following Matlab code:
P = [0.2 0.8; 0.6 0.4];
mu = [0.1 0.9];
while 1
mu_old = mu;
mu = mu * P;
if mu_old == mu
disp(mu);
break;
end
end
Another way of looking at this simple question is that we can see whether the ultimate pmf converges:
Let [math]\displaystyle{ \hat{p_n}(1)=\frac{1}{n}\sum_{k=1}^n I(X_k=1) }[/math] denote the estimator of the stationary probability of state 1,[math]\displaystyle{ \hat{p_n}(2)=\frac{1}{n}\sum_{k=1}^n I(X_k=2) }[/math] denote the estimator of the stationary probability of state 2, where [math]\displaystyle{ \displaystyle I(X_k=1) }[/math] and [math]\displaystyle{ \displaystyle I(X_k=2) }[/math] are indicator variables which equal 1 if [math]\displaystyle{ X_k=1 }[/math](or [math]\displaystyle{ X_k=2 }[/math] for the latter one).
Matlab codes for this explanation is
n=1;
if rand<0.1
x(1)=1;
else
x(1)=0;
end
p1(1)=sum(x)/n;
p2(1)=1-p1(1);
for i=2:10000
n=n+1;
if (x(i-1)==1&rand<0.2)|(x(i-1)==0&rand<0.6)
x(i)=1;
else
x(i)=0;
end
p1(i)=sum(x)/n;
p2(i)=1-p1(i);
end
plot(p1,'red');
hold on;
plot(p2)
The results can be easily seen from the graph below:
Additionally, we can plot the marginal distribution as it converges without estimating it. The following Matlab code shows this:
%transition matrix
P=[0.2 0.8; 0.6 0.4];
%mu at time 0
mu=[0.1 0.9];
%number of points for simulation
n=20;
for i=1:n
mu_a(i)=mu(1);
mu_b(i)=mu(2);
mu=mu*P;
end
t=[1:n];
plot(t, mu_a, t, mu_b);
hleg1=legend('state a', 'state b');
Note that there are chains with stationary distributions that don't converge (the chain might not naturally reach the stationary distribution, and isn't limiting). An example of this is:
[math]\displaystyle{ P = \begin{pmatrix} 0 & 1 & 0 \\ 0 & 0 & 1 \\ 1 & 0 & 0 \end{pmatrix}, \mu_0 = \begin{pmatrix} 1/3 & 1/3 & 1/3 \end{pmatrix} }[/math]
[math]\displaystyle{ \!\mu_0 }[/math] is a stationary distribution, so [math]\displaystyle{ \!\mu P }[/math] is the same for all iterations.
But,
[math]\displaystyle{ P^{1000} = \begin{pmatrix} 0 & 0 & 1 \\ 1 & 0 & 0 \\ 0 & 1 & 0 \end{pmatrix} \ne \begin{pmatrix} \mu \\ \mu \\ \mu \end{pmatrix} }[/math]
So [math]\displaystyle{ \!\mu }[/math] is not a limiting distribution. Also, if
[math]\displaystyle{ \mu = \begin{pmatrix} 0.2 & 0.1 & 0.7 \end{pmatrix} }[/math]
Then [math]\displaystyle{ \!\mu \neq \mu P }[/math].
This can be observed through the following Matlab code.
P = [0 0 1; 1 0 0; 0 1 0]; mu = [0.2 0.1 0.7]; for i= 1:4 mu = mu * P; disp(mu); end
Output:
0.1000 0.7000 0.2000 0.7000 0.2000 0.1000 0.2000 0.1000 0.7000 0.1000 0.7000 0.2000
Note that [math]\displaystyle{ \!\mu_1 = \!\mu_4 }[/math], which indicates that [math]\displaystyle{ \!\mu }[/math] will cycle forever.
This means that this chain has a stationary distribution, but is not limiting. A chain has a limiting distribution iff it is ergodic, that is, aperiodic and positive recurrent. While cycling breaks detailed balance and limiting distribution on the whole state space does not exist, the cycling behavior itself is the "limiting distribution". Also, for each cycles (closed class), we will have a mini-limiting distribution which is useful in analyzing small scale long-term behavior.
Page Rank
Page Rank was the original ranking algorithm used by Google's search engine to rank web pages.<ref> http://ilpubs.stanford.edu:8090/422/ </ref> The algorithm was created by the founders of Google, Larry Page and Sergey Brin as part of Page's PhD thesis. When a query is entered in a search engine, there are a set of web pages which are matched by this query, but this set of pages must be ordered by their "importance" in order to identify the most meaningful results first. Page Rank is an algorithm which assigns importance to every web page based on the links in each page.
Intuition
We can represent web pages by a set of nodes, where web links are represented as edges connecting these nodes. Based on our intuition, there are three main factors in deciding whether a web page is important or not.
- A web page is important if many other pages point to it.
- The more important a webpage is, the more weight is placed on its links.
- The more links a webpage has, the less weight is placed on its links.
Modelling
We can model the set of links as a N-by-N matrix L, where N is the number of web pages we are interested in:
[math]\displaystyle{ L_{ij} = \left\{ \begin{array}{lr} 1 : \text{if page j points to i}\\ 0 : \text{otherwise} \end{array} \right. }[/math]
The number of outgoing links from page j is
[math]\displaystyle{ c_j = \sum_{i=1}^N L_{ij} }[/math]
For example, consider the following set of links between web pages:
According to the factors relating to importance of links, we can consider two possible rankings :
[math]\displaystyle{ \displaystyle 3 \gt 2 \gt 1 \gt 4 }[/math]
or
[math]\displaystyle{ \displaystyle 3\gt 1\gt 2\gt 4 }[/math] if we consider that the high importance of the link from 3 to 1 is more influent than the fact that there are two outgoing links from page 1 and only one from page 2.
We have [math]\displaystyle{ L = \begin{bmatrix}
0 & 0 & 1 & 0 \\
1 & 0 & 0 & 0 \\
1 & 1 & 0 & 1 \\
0 & 0 & 0 & 0
\end{bmatrix} }[/math], and [math]\displaystyle{ c = \begin{pmatrix}2 & 1 & 1 & 1\end{pmatrix} }[/math]
We can represent the ranks of web pages as the vector P, where the ith element is the rank of page i:
[math]\displaystyle{ P_i = (1-d) + d\sum_j \frac{L_{ij}}{c_j} P_j }[/math]
Here we take the sum of the weights of the incoming links, where links are reduced in weight if the linking page has a lot of outgoing links, and links are increased in weight if the linking page has a lot of incoming links.
We don't want to completely ignore pages with no incoming links, which is why we add the constant (1 - d).
If
[math]\displaystyle{ L = \begin{bmatrix} L_{11} & \cdots & L_{1N} \\ \vdots & \ddots & \vdots \\ L_{N1} & \cdots & L_{NN} \end{bmatrix} }[/math]
[math]\displaystyle{ D = \begin{bmatrix} c_1 & \cdots & 0 \\ \vdots & \ddots & \vdots \\ 0 & \cdots & c_N \end{bmatrix} }[/math]
Then [math]\displaystyle{ D^{-1} = \begin{bmatrix} c_1^{-1} & \cdots & 0 \\ \vdots & \ddots & \vdots \\ 0 & \cdots & c_N^{-1} \end{bmatrix} }[/math]
[math]\displaystyle{ \!P = (1-d)e + dLD^{-1}P }[/math]
where [math]\displaystyle{ \!e = \begin{pmatrix} 1 \\ \vdots \\ 1 \end{pmatrix} }[/math] is the vector with all 1's
To simplify the problem, we let [math]\displaystyle{ \!e^T P = N \Rightarrow \frac{e^T P}{N} = 1 }[/math]. This means that the average importance of all pages on the internet is 1.
Then [math]\displaystyle{ \!P = (1-d)\frac{ee^TP}{N} + dLD^{-1}P }[/math]
- [math]\displaystyle{ \! = \left [ (1-d)\frac{ee^T}{N} + dLD^{-1} \right ] P }[/math]
- [math]\displaystyle{ \! = \left [ \left ( \frac{1-d}{N} \right ) E + dLD^{-1} \right ] P }[/math], where [math]\displaystyle{ E }[/math] is an NxN matrix filled with ones.
Let [math]\displaystyle{ \!A = \left [ \left ( \frac{1-d}{N} \right ) E + dLD^{-1} \right ] }[/math]
Then [math]\displaystyle{ \!P = AP }[/math].
Note that P is a stationary distribution and, more importantly, P is an eigenvector of A with eigenvalue 1. Therefore, we can find the ranks of all web pages by solving this equation for P.
We can find the vector P for the example above, using the following Matlab code:
L = [0 0 1 0; 1 0 0 0; 1 1 0 1; 0 0 0 0]; D = [2 0 0 0; 0 1 0 0; 0 0 1 0; 0 0 0 1]; d = 0.8 ;% pages with no links get a weight of 0.2 N = 4 ;
A = ((1-d)/N) * ones(N) + d * L * inv(D); [EigenVectors, EigenValues] = eigs(A) s=sum(EigenVectors(:,1));% we should note that the average entry of P should be 1 according to our assumption P=(EigenVectors(:,1))/s*N
This outputs:
EigenVectors = -0.6363 0.7071 0.7071 -0.0000 -0.3421 -0.3536 + 0.3536i -0.3536 - 0.3536i -0.7071 -0.6859 -0.3536 - 0.3536i -0.3536 + 0.3536i 0.0000 -0.0876 0.0000 + 0.0000i 0.0000 - 0.0000i 0.7071
EigenValues =
1.0000 0 0 0
0 -0.4000 - 0.4000i 0 0
0 0 -0.4000 + 0.4000i 0
0 0 0 0.0000
P =
1.4528 0.7811 1.5660 0.2000
Note that there is an eigenvector with eigenvalue 1. The reason why there always exist a 1-eigenvector is that A is a stochastic matrix.
Thus our vector P is [math]\displaystyle{ \begin{bmatrix}1.4528 \\ 0.7811 \\ 1.5660\\ 0.2000 \end{bmatrix} }[/math]
However, this method is not practical, because there are simply too many web pages on the internet. So instead Google uses a method to approximate an eigenvector with eigenvalue 1.
Note that page three has the rank with highest magnitude and page four has the rank with lowest magnitude, as expected.
Markov Chain Monte Carlo - Metropolis-Hastings - October 25th, 2011
We want to find [math]\displaystyle{ \int h(x)f(x)\, \mathrm dx }[/math], but we don't know how to sample from [math]\displaystyle{ \,f }[/math].
We have seen simple techniques earlier in the course; here is an example of a real life application. It consists of the search for a Markov Chain such that its stationary distribution is [math]\displaystyle{ \,f }[/math].
Main procedure
Let us suppose that [math]\displaystyle{ \,q(y|x) }[/math] is a friendly distribution: we can sample from this function.
1. Initialize the chain with [math]\displaystyle{ \,x_{i} }[/math] and set [math]\displaystyle{ \,i=0 }[/math].
2. Draw a point from [math]\displaystyle{ \,q(y|x) }[/math] i.e. [math]\displaystyle{ \,Y \backsim q(y|x_{i}) }[/math].
3. Evaluate [math]\displaystyle{ \,r(x,y)=min\left\{\frac{f(y)}{f(x)}\frac{q(x|y)}{q(y|x)},1\right\} }[/math]
4. Draw a point [math]\displaystyle{ \,U \backsim Unif[0,1] }[/math].
5. [math]\displaystyle{ \,x_{i+1}=\begin{cases}y & \text{ if } U\lt r \\x_{i} & \text{ otherwise } \end{cases} }[/math].
6. [math]\displaystyle{ \,i=i+1 }[/math]. Go back to 2.
Remark 1
A very common choice for [math]\displaystyle{ \,q(y|x) }[/math] is [math]\displaystyle{ \,N(y;x,b^{2}) }[/math], a normal distribution centered at the current point.
Note : In this case [math]\displaystyle{ \,q(y|x) }[/math] is symmetric i.e. [math]\displaystyle{ \,q(y|x)=q(x|y) }[/math].
(Because [math]\displaystyle{ \,q(y|x)=\frac{1}{\sqrt{2\pi}b}e^{-\frac{1}{2b^{2}}(y-x)^{2}} }[/math] and [math]\displaystyle{ \,(y-x)^{2}=(x-y)^{2} }[/math]).
Thus we have [math]\displaystyle{ \,\frac{q(x|y)}{q(y|x)}=1 }[/math], which implies :
[math]\displaystyle{ \,r(x,y)=min\left\{\frac{f(y)}{f(x)},1\right\} }[/math].
In general, if [math]\displaystyle{ \,q(x|y) }[/math] is symmetric then the algorithm is called Metropolis in reference to the original algorithm (made in 1953)<ref>http://en.wikipedia.org/wiki/Equations_of_State_Calculations_by_Fast_Computing_Machines</ref>.
Remark 2
The value y is accepted if [math]\displaystyle{ \,u\lt min\left\{\frac{f(y)}{f(x)},1\right\} }[/math] so it is accepted with the probability [math]\displaystyle{ \,min\left\{\frac{f(y)}{f(x)},1\right\} }[/math].
Thus, if [math]\displaystyle{ \,f(y)\gt f(x) }[/math], then [math]\displaystyle{ \,y }[/math] is always accepted.
The higher that value of the pdf is in the vicinity of a point [math]\displaystyle{ \,y_1 }[/math], the more likely it is that a random variable will take on values around [math]\displaystyle{ \,y_1 }[/math]. As a result it makes sense that we would want a high probability of acceptance for points generated near [math]\displaystyle{ \,y_1 }[/math].
Remark 3
One strength of the Metropolis-Hastings algorithm is that normalizing constants, which are often quite difficult to determine, can be cancelled out in the ratio [math]\displaystyle{ r }[/math]. For example, consider the case where we want to sample from the beta distribution, which has the pdf:
[math]\displaystyle{ \begin{align} f(x;\alpha,\beta)& = \frac{1}{\mathrm{B}(\alpha,\beta)}\, x^{\alpha-1}(1-x)^{\beta-1}\end{align} }[/math]
The beta function, B, appears as a normalizing constant but it can be simplified by construction of the method.
Example
[math]\displaystyle{ \,f(x)=\frac{1}{\pi^{2}}\frac{1}{1+x^{2}} }[/math]
Then, we have [math]\displaystyle{ \,f(x)\propto\frac{1}{1+x^{2}} }[/math].
And let us take [math]\displaystyle{ \,q(x|y)=\frac{1}{\sqrt{2\pi}b}e^{-\frac{1}{2b^{2}}(y-x)^{2}} }[/math].
Then [math]\displaystyle{ \,q(x|y) }[/math] is symmetric.
Therefore Y can be simplified.
We get :
[math]\displaystyle{ \,\begin{align} \displaystyle r(x,y) & =min\left\{\frac{f(y)}{f(x)}\frac{q(x|y)}{q(y|x)},1\right\} \\ & =min\left\{\frac{f(y)}{f(x)},1\right\} \\ & =min\left\{ \frac{ \frac{1}{1+y^{2}} }{ \frac{1}{1+x^{2}} },1\right\}\\ & =min\left\{ \frac{1+x^{2}}{1+y^{2}},1\right\}\\ \end{align} }[/math].
The Matlab code of the algorithm is the following :
clear all
close all
clc
b=2;
x(1)=randn;
for i=2:10000
y=b*randn+x(i-1);
r=min((1+x(i-1)^2)/(1+y^2),1);
u=rand;
if u<r
x(i)=y;
else
x(i)=x(i-1);
end
end
hist(x(5000:end));
%The Markov Chain usually takes some time to converge and this is known as the "burning time".
%Therefore, we don't display the first 5000 points because they don't show the limiting behaviour of the Markov
Chain.
As we can see, the choice of the value of b is made by us.
Changing this value has a significant impact on the results we obtain. There is a pitfall when b is too big or too small.
Example with [math]\displaystyle{ \,b=0.1 }[/math] (Also with graph after we run j=5000:10000; plot(j,x(5000:10000)):
With [math]\displaystyle{ \,b=0.1 }[/math], the chain takes small steps so the chain doesn't explore enough of the sample space. It doesn't give an accurate report of the function we want to sample.
Example with [math]\displaystyle{ \,b=10 }[/math] :
With [math]\displaystyle{ \,b=10 }[/math], jumps are very unlikely to be accepted as they deviate far from the mean (i.e. [math]\displaystyle{ \,y }[/math] is rejected as [math]\displaystyle{ \ u\gt r }[/math] and [math]\displaystyle{ \,x(i)=x(i-1) }[/math] most of the time, hence most sample points stay fairly close to the origin. The third graph that resembles white noise (as in the case of [math]\displaystyle{ \,b=2 }[/math]) indicates better sampling as more points are covered and accepted. For [math]\displaystyle{ \,b=0.1 }[/math], we have lots of jumps but most values are not repeated, hence the stationary distribution is less obvious; whereas in the [math]\displaystyle{ \,b=10 }[/math] case, many points remains around 0. Approximately 73% were selected as x(i-1).
Example with [math]\displaystyle{ \,b=2 }[/math] :
With [math]\displaystyle{ \,b=2 }[/math], we get a more accurate result as we avoid these extremes. Approximately 37% were selected as x(i-1).
If the sample from the Markov Chain starts to look like the target distribution quickly, we say the chain is mixing well.
Theory and Applications of Metropolis-Hastings - October 27th, 2011
As mentioned in the previous section, the idea of the Metropolis-Hastings (MH) algorithm is to produce a Markov chain that converges to a stationary distribution [math]\displaystyle{ \!f }[/math] which we are interested in sampling from.
Convergence
One important fact to check is that [math]\displaystyle{ \displaystyle f }[/math] is indeed a stationary distribution in the MH scheme. For this, we can appeal to the implications of the detailed balance property:
Given a probability vector [math]\displaystyle{ \!\pi }[/math] and a transition matrix [math]\displaystyle{ \displaystyle P }[/math], [math]\displaystyle{ \!\pi }[/math] has the detailed balance property if [math]\displaystyle{ \!\pi_iP_{ij} = P_{ji}\pi_j }[/math]
If [math]\displaystyle{ \!\pi }[/math] satisfies detailed balance, then it is a stationary distribution.
The above definition applies to the case where the states are discrete. In the continuous case, [math]\displaystyle{ \displaystyle f }[/math] satisfies detailed balance if [math]\displaystyle{ \displaystyle f(x)p(x,y)=f(y)p(y,x) }[/math]. Where [math]\displaystyle{ \displaystyle p(x,y) }[/math] and [math]\displaystyle{ \displaystyle p(y,x) }[/math] are the probabilities of transitioning from x to y and y to x respectively. If we can show that [math]\displaystyle{ \displaystyle f }[/math] has the detailed balance property, we can conclude that it is a stationary distribution. Because [math]\displaystyle{ \int^{}_y f(y)p(y,x)dy=\int^{}_y f(x)p(x,y)dy=f(x) }[/math].
In the MH algorithm, we use a proposal distribution to generate [math]\displaystyle{ \, y }[/math]~[math]\displaystyle{ \displaystyle q(y|x) }[/math], and accept y with probability [math]\displaystyle{ min\left\{\frac{f(y)}{f(x)}\frac{q(x|y)}{q(y|x)},1\right\} }[/math]
Suppose, without loss of generality, that [math]\displaystyle{ \frac{f(y)}{f(x)}\frac{q(x|y)}{q(y|x)} \le 1 }[/math]. This implies that [math]\displaystyle{ \frac{f(x)}{f(y)}\frac{q(y|x)}{q(x|y)} \ge 1 }[/math]
Let [math]\displaystyle{ \,r(x,y) }[/math] be the chance of accepting point y given that we are at point x.
So [math]\displaystyle{ \,r(x,y) = min\left\{\frac{f(y)}{f(x)}\frac{q(x|y)}{q(y|x)},1\right\} = \frac{f(y)}{f(x)} \frac{q(x|y)}{q(y|x)} }[/math]
Let [math]\displaystyle{ \,r(y,x) }[/math] be the chance of accepting point x given that we are at point y.
So [math]\displaystyle{ \,r(y,x) = min\left\{\frac{f(x)}{f(y)}\frac{q(y|x)}{q(x|y)},1\right\} = 1 }[/math]
[math]\displaystyle{ \,p(x,y) }[/math] is the probability of generating and accepting y, while at point x.
So [math]\displaystyle{ \,p(x,y) = q(y|x)r(x,y) = q(y|x) \frac{f(y)}{f(x)} \frac{q(x|y)}{q(y|x)} = \frac{f(y)q(x|y)}{f(x)} }[/math]
[math]\displaystyle{ \,p(y,x) }[/math] is the probability of generating and accepting x, while at point y.
So [math]\displaystyle{ \,p(y,x) = q(x|y)r(y,x) = q(x|y) }[/math]
[math]\displaystyle{ \,f(x)p(x,y) = f(x)\frac{f(y)q(x|y)}{f(x)} = f(y)q(x|y) = f(y)p(y,x) }[/math]
Thus, detailed balance holds.
- i.e. [math]\displaystyle{ \,f(x) }[/math] is stationary distribution
It can be shown (although not here) that [math]\displaystyle{ \!f }[/math] is a limiting distribution as well. Therefore, the MH algorithm generates a sequence whose distribution converges to [math]\displaystyle{ \!f }[/math], the target.
Implementation
In the implementation of MH, the proposal distribution is commonly chosen to be symmetric, which simplifies the calculations and makes the algorithm more intuitively understandable. The MH algorithm can usually be regarded as a random walk along the distribution we want to sample from. Suppose we have a distribution [math]\displaystyle{ \!f }[/math]:
Suppose we start the walk at point [math]\displaystyle{ \!x }[/math]. The point [math]\displaystyle{ \!y_{1} }[/math] is in a denser region than [math]\displaystyle{ \!x }[/math], therefore, the walk will always progress from [math]\displaystyle{ \!x }[/math] to [math]\displaystyle{ \!y_{1} }[/math]. On the other hand, [math]\displaystyle{ \!y_{2} }[/math] is in a less dense region, so it is not certain that the walk will progress from [math]\displaystyle{ \!x }[/math] to [math]\displaystyle{ \!y_{2} }[/math]. In terms of the MH algorithm:
[math]\displaystyle{ r(x,y_{1})=min(\frac{f(y_{1})}{f(x)},1)=1 }[/math] since [math]\displaystyle{ \!f(y_{1})\gt f(x) }[/math]. Thus, any generated value with a higher density will be accepted.
[math]\displaystyle{ r(x,y_{2})=\frac{f(y_{2})}{f(x)} }[/math]. The lower the density of [math]\displaystyle{ \!y_{2} }[/math] is, the less chance it will have of being accepted.
A certain class of proposal distributions can be written in the form:
[math]\displaystyle{ \,y|x_i = x_i + \epsilon_i }[/math]
where [math]\displaystyle{ \,\epsilon_i = g(|x-y|) }[/math]
The density depends only on the distance between the current point and the next one (which can be seen as the "step" being taken). These proposal distributions give the Markov chain the random walk nature. The normal distribution that we frequently use in our examples satisfies the above definition.
In actual implementations of the MH algorithm, the proposal distribution needs to be chosen judiciously, because not all proposals will work well with all target distributions we want to sample from. Take a trimodal distribution for example:
If we choose the proposal distribution to be a standard normal as we have done before, problems will arise. The low densities between the peaks means that the MH algorithm will almost never walk to any points generated in these regions and get stuck at one peak. One way to address this issue is to increase the variance, so that the steps will be large enough to cross the gaps. Of course, in this case, it would probably be beneficial to come up with a different proposal function. As a rule of thumb, such functions should result in an approximately 50% acceptance rate for generated points.
Simulated Annealing
Metropolis-Hastings is very useful in simulation methods for solving optimization problems. One such application is simulated annealing, which addresses the problems of minimizing a function [math]\displaystyle{ \!h(x) }[/math]. This method will not always produce the global solution, but it is intuitively simple and easy to implement.
Consider [math]\displaystyle{ e^{\frac{-h(x)}{T}} }[/math], maximizing this expression is equivalent to minimizing [math]\displaystyle{ \!h(x) }[/math]. Suppose [math]\displaystyle{ \mu }[/math] is the maximizing value and [math]\displaystyle{ h(x)=(x-\mu)^2 }[/math], then the maximization function is a gaussian distribution [math]\displaystyle{ \!e^{-\frac{(x-\mu)^2}{T}} }[/math]. When many samples are taken from this distribution, the mean will converge to the desired maximizing value. The annealing comes into play by lowering T (the temperature) as the sampling progresses, making the distribution narrower. The steps of simulated annealing are outlined below:
1. start with a random [math]\displaystyle{ \!x }[/math] and set T to a large number
2. generate [math]\displaystyle{ \!y }[/math] from a proposal distribution [math]\displaystyle{ \!q(y|x) }[/math], which should be symmetric
3. accept [math]\displaystyle{ \!y }[/math] with probability [math]\displaystyle{ min(\frac{f(y)}{f(x)},1) }[/math] . (Note: [math]\displaystyle{ f(x) = e^{\frac{-h(x)}{T}} }[/math])
4. decrease T, and then go to step 2
To decrease T in Step 4, a variety of functions can be used. For example, a common temperature function used is with geometric decline, given by an initial temperature [math]\displaystyle{ \! T_o }[/math], final temperature [math]\displaystyle{ \! T_f }[/math], the number of steps n, and the temperature function [math]\displaystyle{ \ T(t) = T_0(\frac{T_f}{T_o})^{t/n} }[/math]
The following plot and Matlab code illustrates the simulated annealing procedure as temperature T, the variance, decreases for a Gaussian distribution with zero mean. Starting off with a large value for the temperature T allows the Metropolis-Hastings component of the procedure to capture the mean, before gradually decreasing the temperature T in order to converge to the mean.
x=-10:0.1:10;
mu=0;
T=5;
colour = ['b', 'g', 'm', 'r', 'k'];
for i=1:5
pdfNormal=normpdf(x, mu, T);
plot(x, pdfNormal, colour(i));
T=T-1;
hold on
end
hleg1=legend('T=5', 'T=4', 'T=3', 'T=2', 'T=1');
title('Simulated Annealing Illustration');
References
<references/>
Simulated Annealing and Gibbs Sampling - November 1, 2011
continued from previous lecture...
Recall [math]\displaystyle{ \ r(x,y)=min(\frac{f(y)}{f(x)},1) }[/math] where [math]\displaystyle{ \frac{f(y)}{f(x)} = \frac{e^{\frac{-h(y)}{T}}}{e^{\frac{-h(x)}{T}}} = e^{\frac{h(x)-h(y)}{T}} }[/math] where [math]\displaystyle{ \ r(x,y) }[/math] represents the probability of accepting [math]\displaystyle{ \ y }[/math].
We will now look at a couple cases where [math]\displaystyle{ \displaystyle h(y) \gt h(x) }[/math] or [math]\displaystyle{ \displaystyle h(y) \lt h(x) }[/math], and explore whether to accept or reject [math]\displaystyle{ \ y }[/math].
Cases
Case a) Suppose [math]\displaystyle{ \displaystyle h(y) \lt h(x) }[/math]. Since we want to find the minimum value for [math]\displaystyle{ \displaystyle h(x) }[/math], and the point [math]\displaystyle{ \displaystyle y }[/math] creates a lower value than our previous point, we accept the new point. Mathematically, [math]\displaystyle{ \displaystyle h(y) \lt h(x) }[/math] implies that:
[math]\displaystyle{ \frac{f(y)}{f(x)} \gt 1 }[/math]. Therefore, [math]\displaystyle{ \displaystyle r = 1 }[/math]. So, we will always accept [math]\displaystyle{ \displaystyle y }[/math].
Case b) Suppose [math]\displaystyle{ \displaystyle h(y) \gt h(x) }[/math]. This is bad, since our goal is to minimize [math]\displaystyle{ \displaystyle h(x) }[/math]. However, we may still accept [math]\displaystyle{ \displaystyle y }[/math] with some chance:
[math]\displaystyle{ \frac{f(y)}{f(x)} \lt 1 }[/math]. Therefore, [math]\displaystyle{ \displaystyle r \lt 1 }[/math]. So, we may accept [math]\displaystyle{ \displaystyle y }[/math] with probability [math]\displaystyle{ \displaystyle r }[/math].
Next, we will look at these cases as [math]\displaystyle{ \displaystyle T\to0 }[/math].
As [math]\displaystyle{ \displaystyle T\to0 }[/math] and case a) happens, [math]\displaystyle{ e^{\frac{h(x)-h(y)}{T}} \to \infty }[/math], so we will always accept [math]\displaystyle{ \displaystyle y }[/math].
As [math]\displaystyle{ \displaystyle T\to0 }[/math] and case b) happens, [math]\displaystyle{ e^{\frac{h(x)-h(y)}{T}} \to \ 0 }[/math], so the probability that [math]\displaystyle{ \displaystyle y }[/math] will be accepted gets extremely small.
It is worth noting that if we simply start with a small value of T, we may end up rejecting all the generated points, and hence we will get stuck somewhere in the function (due to case b)), which might be a maximum value in some intervals (local maxima), but not in the whole domain (global maxima). It is therefore necessary to start with a large value of T in order to explore the whole function. At the same time, a good estimation of [math]\displaystyle{ \ x_0 }[/math] < is needed (at least cannot differ from the maximum point too much).
Example
Let [math]\displaystyle{ \displaystyle h(x) = (x-2)^2 }[/math]. The graph of it is:
Then, [math]\displaystyle{ e^{\frac{-h(x)}{T}} = e^{\frac{-(x-2)^2}{T}} }[/math] . Take an initial value of T = 20. A graph of this is:
In comparison, we look a graph of T = 0.2:
One can see that with a low T value, the graph has a lot of r = 0, and r >1, while having a bigger T value gives smoother transitions in the graph.
The MATLAB code for the above graphs are:
ezplot('(x-2)^2',[-6,10])
ezplot('exp((-(x-2)^2)/20)',[-6,10])
ezplot('exp((-(x-2)^2)/0.2)',[-6,10])
Travelling Salesman Problem
The simulated annealing method can be applied to compute the solution to the travelling salesman problem. Suppose there are N cities and the salesman only have to visit each city once. The objective is to find out the shortest path (i.e. shortest total length of journey) connecting the cities. An algorithm using simulated annealing on the problem can be found here (Reference).
Gibbs Sampling
Gibbs sampling is another Markov chain Monte Carlo method, similar to Metropolis-Hastings. There are two main differences between Metropolis-Hastings and Gibbs sampling. First, the candidate state is always accepted as the next state in Gibbs sampling. Second, it is assumed that the full conditional distributions are known, i.e. [math]\displaystyle{ P(X_i=x|X_j=x_j, \forall j\neq i) }[/math] for all [math]\displaystyle{ \displaystyle i }[/math]. The idea is that it is easier to sample from conditional distributions which are sets of one dimensional distributions than to sample from a joint distribution which is a higher dimensional distribution. Gibbs is a way to turn the joint distribution into multiple conditional distributions.
Advantages:
- Sampling from conditional distributions may be easier than sampling from joint distributions
Disadvantages:
- We do not necessarily know the conditional distributions
For example, if we want to sample from [math]\displaystyle{ \, f_{X,Y}(x,y) }[/math], we need to know how to sample from [math]\displaystyle{ \, f_{X|Y}(x|y) }[/math] and [math]\displaystyle{ \, f_{Y|X}(y|x) }[/math]. Suppose the chain starts with [math]\displaystyle{ \,(X_0,Y_0) }[/math] and [math]\displaystyle{ (X_1,Y_1), \dots , (X_n,Y_n) }[/math] have been sampled. Then,
[math]\displaystyle{ \, (X_{n+1},Y_{n+1})=(f_{X|Y}(x|Y_n),f_{Y|X}(y|X_{n+1})) }[/math]
Gibbs sampling turns a multi-dimensional distribution into a set of one-dimensional distributions. If we want to sample from
[math]\displaystyle{ P_{X^1,\dots ,X^p}(x^1,\dots ,x^p) }[/math]
and the full conditionals are known, then:
[math]\displaystyle{ X^1_{n+1}=f(X^1|X^2_n,\dots ,X^p_n) }[/math]
[math]\displaystyle{ X^2_{n+1}=f(X^2|X^1_{n+1},X^3_n\dots ,X^p_n) }[/math]
[math]\displaystyle{ \vdots }[/math]
[math]\displaystyle{ X^{p-1}_{n+1}=f(X^{p-1}|X^1_{n+1},\dots ,X^{p-2}_{n+1},X^p_n) }[/math]
[math]\displaystyle{ X^p_{n+1}=f(X^p|X^1_{n+1},\dots ,X^{p-1}_{n+1}) }[/math]
With Gibbs sampling, we can simulate [math]\displaystyle{ \displaystyle n }[/math] random variables sequentially from [math]\displaystyle{ \displaystyle n }[/math] univariate conditionals rather than generating one [math]\displaystyle{ \, n }[/math]-dimensional vector using the full joint distribution, which could be a lot more complicated.
Computational inference deals with probabilistic graphical models. Gibbs sampling is useful here: graphical models show the dependence relations among random variables. For instance, Bayesian networks are graphical models represented using directed acyclic graphs. Looking at such a graphical model tells us on which random variable the distribution of a certain random variable depends (i.e. its parent). The model can be used to "factor" a joint distribution into conditional distributions.
For example, consider the five random variables A, B, C, D, and E. Without making any assumptions about dependence relations among them, all we know is
[math]\displaystyle{ \, P(A,B,C,D,E)= }[/math][math]\displaystyle{ \, P(A|B,C,D,E) P(B|C,D,E) P(C|D,E) P(D|E) P(E) }[/math]
However, if we know the relation between the random variables, e.g. given the graphical model on the left, we can simplify this expression:
[math]\displaystyle{ \, P(A,B,C,D,E)=P(A) P(B|A) P(C|A) P(D|C) P(E|C) }[/math]
Although the joint distribution may be very complicated, the conditional distributions may not be.
Check out the following notes on Gibbs sampling:
- MCMC and Gibbs Sampling, MIT Lecture Notes
- chapter 7.4 in Notes on Computational Methods in Statistics
- chapter 4.9 in Introduction to Probability Models by Sheldon Ross
Example of Gibbs sampling: Multi-variate normal
We'd like to generate samples from a bivariate normal with parameters
[math]\displaystyle{ \mu = \begin{bmatrix}1\\ 2 \end{bmatrix} = \begin{bmatrix}\mu_1 \\ \mu_2 \end{bmatrix} }[/math] and [math]\displaystyle{ \sigma = \begin{bmatrix}1 && 0.9 \\ 0.9 && 1 \end{bmatrix}= \begin{bmatrix}1 && \rho \\ \rho && 1 \end{bmatrix} }[/math]
The conditional distributions of multi-variate normal random variables are also normal:
[math]\displaystyle{ \, f(x_1|x_2)=N(\mu_1 + \rho(x_2-\mu_2), 1-\rho^2) }[/math]
[math]\displaystyle{ \, f(x_2|x_1)=N(\mu_2 + \rho(x_1-\mu_1), 1-\rho^2) }[/math]
In general, if the joint distribution has parameters
[math]\displaystyle{ \mu = \begin{bmatrix}\mu_1 \\ \mu_2 \end{bmatrix} }[/math] and [math]\displaystyle{ \Sigma = \begin{bmatrix} \Sigma _{1,1} && \Sigma _{1,2} \\ \Sigma _{2,1} && \Sigma _{2,2} \end{bmatrix} }[/math]
then the conditional distribution [math]\displaystyle{ \, f(x_1|x_2) }[/math] has mean [math]\displaystyle{ \, \mu_{1|2} = \mu_1 + \Sigma _{1,2}(\Sigma _{2,2})^{-1}(x_2-\mu_2) }[/math] and variance [math]\displaystyle{ \, \Sigma _{1|2} = \Sigma _{1,1}-\Sigma _{1,2}(\Sigma _{2,2})^{-1}\Sigma _{2,1} }[/math].
Thus, the algorithm for Gibbs sampling is:
- 1) Set i = 1
- 2) Draw [math]\displaystyle{ X_1^{(i)} }[/math] ~ [math]\displaystyle{ N(\mu_1 + \rho(X_2^{(i-1)}-\mu_2), 1-\rho^2) }[/math]
- 3) Draw [math]\displaystyle{ X_2^{(i)} }[/math] ~ [math]\displaystyle{ N(\mu_2 + \rho(X_1^{(i)}-\mu_1), 1-\rho^2) }[/math]
- 4) Set [math]\displaystyle{ X^{(i)} = [X_1^{(i)}, X_2^{(i)}]^T }[/math]
- 5) Increment i, return to step 2.
The Matlab code implementation of this algorithm is as follows:
mu = [1; 2];
sigma = [1 0.9; 0.9 1];
X(:,1) = [1; 2];
r = 1 - 0.9^2;
for i = 2:2000
X(1,i) = 1 + 0.9*(X(2,i-1) - mu(2)) + r*randn;
X(2,i) = 2 + 0.9*(X(1,i) - mu(1)) + r*randn;
end
plot(X(1,:),X(2,:),'.')
Which gives the following plot:
Principal Component Analysis (PCA) - November 8, 2011
Principal Component Analysis is a 100 year old algorithm used for reducing the dimensionality of data. As the number of dimensions increase, the number of data points needed to sample accurately increase by an exponential factor.
[math]\displaystyle{ \, x\in \mathbb{R}^D \rarr y\in \mathbb{R}^d }[/math]
[math]\displaystyle{ \ d \le D }[/math]
We want to transform [math]\displaystyle{ \, x }[/math] to [math]\displaystyle{ \, y }[/math] such that we reduce the dimensionality yet lose little information. Generally, variation in the data provides information. Thus we would like to reduce the dimensionality but keep as much variation, or information, as the original set of data. Also, covariance amongst the data reduces the amount of information we can infer from the data. Therefore, we would like to also reduce covariance when we reduce dimensionality.
For example, consider dots in a three dimensional space. By unrolling the 2D manifold that they are on, we can reduce the data to 2D while losing little information. Note: This is not an application of PCA, but it simply illustrates one way we can reduce dimensionality.
Principle Component Analysis allows us to reduce data to a linear subspace of its original space. It works best when data is in a lower dimensional subspace of its original space.
Probabilistic View
We can see a data set [math]\displaystyle{ \, x }[/math] as a high dimensional random variable governed by a low dimensional random variable [math]\displaystyle{ \, y }[/math]. Given [math]\displaystyle{ \, x }[/math], we are trying to estimate [math]\displaystyle{ \, y }[/math].
We can see this in 2D linear regression, as the locations of data points in a scatter plot are governed by its approximate linear regression. The subspace that we have reduced the data to here is in the direction of variation in the data.
Principal Component Analysis
Principal component analysis is an orthogonal linear transformation on a data set. It associates the data coordinates with a new set of orthogonal vectors, each representing the direction of the maximum variance of the data. That is, the first principal component is the direction of the maximum variance, the second principal component is the direction of the maximum variance orthogonal to the first principal component, the third principal component is the direction of the maximum variance orthogonal to the first and second principal component and so on, until we have D principal components, where D is the dimension of the original data.
Suppose we have data represented by [math]\displaystyle{ \, X \in \mathbb{R}^{D \times n} }[/math]
Note that we are assuming that the data is mean centered. In other words, the average of every row is zero. If it isn't, we shift the data to have a mean of zero by subtracting the mean of every row from each [math]\displaystyle{ \ x }[/math] value in that row. This pre-processing step essentially alters the data such that each row in [math]\displaystyle{ \ X }[/math] includes only how the data differs from the mean of the sample, hence ensuring that the first PC describes the direction of maximum variance.
To find the first principal component, we want to find a unit vector [math]\displaystyle{ \ W \in \mathbb{R}^{D} }[/math] that maximizes the variance of [math]\displaystyle{ \,W^TX }[/math]. We restrict [math]\displaystyle{ \,W }[/math] to unit vectors since we are only looking for the direction of the vector of maximum variation: the actual scale of it is unnecessary. So [math]\displaystyle{ \,W^TW = 1 }[/math].
The variance of [math]\displaystyle{ \,W^TX }[/math] is [math]\displaystyle{ \,W^TSW }[/math] where [math]\displaystyle{ \,S }[/math] is the covariance matrix of X.
[math]\displaystyle{ \, S = (X-\mu)(X-\mu)^T = XX^T }[/math], since [math]\displaystyle{ \ \mu }[/math] is just the zero vector after we center the data around the mean.
So we have to solve the problem
[math]\displaystyle{ \, \text {Max } W^TSW \text{ such that } W^TW = 1 }[/math]
Using the method of Lagrange multipliers, we have
[math]\displaystyle{ \,L(W, \lambda) = W^TSW - \lambda(W^TW - 1) }[/math]
We set
[math]\displaystyle{ \, \frac{\partial L}{\partial W} = 0 }[/math]
Note that [math]\displaystyle{ \, W^TSW }[/math] is a quadratic form. So we have
[math]\displaystyle{ \, \frac{\partial L}{\partial W} = 2SW - 2\lambda W = 0 }[/math]
[math]\displaystyle{ \, SW = \lambda W }[/math]
Since S is a matrix and lambda is a scaler, W is an eigenvector of S and lambda is its corresponding eigenvalue.
Suppose that
[math]\displaystyle{ \, \lambda_1 \ge \lambda_2 \ge \cdots \ge \lambda_D }[/math] are eigenvalues of S and [math]\displaystyle{ \, u_1, u_2, \cdots u_D }[/math] are their corresponding eigenvectors.
We want to choose some [math]\displaystyle{ \, W = u }[/math]
[math]\displaystyle{ \,u^TSu =u^T\lambda u = \lambda u^Tu = \lambda }[/math]
So to maximize [math]\displaystyle{ \, u^TSu }[/math], choose the eigenvector corresponding to the max eiegenvalue, e.g. [math]\displaystyle{ \, u_1 }[/math].
So we let [math]\displaystyle{ \, W = u_1 }[/math] be the first principal component.
The principal components decompose the total variance in the data.
[math]\displaystyle{ \, \sum_{i=1}^D \text{Var}(u_i) = \sum_{i=1}^D \lambda_i = \text{Tr}(S) = \sum_{i=1}^D \text{Var}(x_i) }[/math]
Singular Value Decomposition
Singular value decomposition is a "generalization" of eigenvalue decomposition "to rectangular matrices of size mxn."<ref name="Abdel_SVD">Abdel-Rahman, E. (2011). Singular Value Decomposition [Lecture notes]. Retrieved from http://uwace.uwaterloo.ca</ref> Singular value decomposition solves:
- [math]\displaystyle{ \ A_{m\times n}\ v_{n\times 1}=s\ u_{m\times 1} }[/math]
"for the right singular vector v, the singular value s, and the left singular vector u. There are n singular values si and n right and left singular vectors that must satisfy the following conditions"<ref name="Abdel_SVD"/>:
- "All singular values are non-negative"<ref name="Abdel_SVD"/>,
[math]\displaystyle{ \ s_i \ge 0. }[/math] - All "right singular vectors are pairwise orthonormal"<ref name="Abdel_SVD"/>,
[math]\displaystyle{ \ v_i^{T}v_j=\delta_{i,j}. }[/math] - All "left singular vectors are pairwise orthonormal"<ref name="Abdel_SVD"/>,
[math]\displaystyle{ \ u_i^{T}u_j=\delta_{i,j}. }[/math]
where
- [math]\displaystyle{ \delta_{i,j}=\left\{\begin{matrix}1 & \mathrm{if}\ i=j \\ 0 & \mathrm{if}\ i\neq j\end{matrix}\right. }[/math]
Procedure to find the singular values and vectors
Observe the following about the eigenvalue decomposition of a real square matrix A where v is the unit eigenvector:
- [math]\displaystyle{ \begin{align} & Av=\lambda v \\ & (Av)^T=(\lambda v)^T \\ & (Av)^TAv=(\lambda v)^T\lambda v \\ & v^TA^TAv=\lambda^2v^Tv \\ & vv^TA^TAv=v\lambda^2 \\ & A^TAv=\lambda^2v \end{align} }[/math]
As a result:
- "The matrices A and ATA have the same eigenvectors."<ref name="Abdel_SVD"/>
- "The eigenvalues of matrix ATA are the square of the eigenvalues of matrix A."<ref name="Abdel_SVD"/>
- Since matrix ATA is symmetric for any matrix A,
- "all the eigenvalues of matrix ATA are real and distinct."<ref name="Abdel_SVD"/>
- "the eigenvectors of matrix ATA are orthogonal and can be chosen to be orthonormal."<ref name="Abdel_SVD"/>
- "The eigenvalues of matrix ATA are non-negative"<ref name="Abdel_SVD"/> since [math]\displaystyle{ \ \lambda^2_i \ge 0. }[/math]
Conclusions 3 and 4 are "true even for a rectangular matrix A since ATA is still a square symmetric matrix"<ref name="Abdel_SVD"/> and its eigenvalues and eigenvectors can be found.
Therefore, for a rectangular matrix A, assuming m>n, the singular values and vectors can be found by:
- "Form the nxn symmetric matrix ATA."<ref name="Abdel_SVD"/>
- Perform an eigenvalue decomposition to get n eigenvalues and their "corresponding eigenvectors, ordered such that"<ref name="Abdel_SVD"/>
[math]\displaystyle{ \lambda_1 \ge \lambda_2 \ge \dots \ge \lambda_n \ge 0 }[/math] and [math]\displaystyle{ \{v_1, v_2, \dots, v_n\}. }[/math] - "The singular values are"<ref name="Abdel_SVD"/>:
[math]\displaystyle{ s_1=\sqrt{\lambda_1} \ge s_2=\sqrt{\lambda_2} \ge \dots \ge s_n=\sqrt{\lambda_n} \ge 0. }[/math]
"The non-zero singular values are distinct; the equal sign applies only to the singular values that are equal to zero."<ref name="Abdel_SVD"/> - "The n-dimensional right singular vectors are"<ref name="Abdel_SVD"/>
[math]\displaystyle{ \{v_1, v_2, \dots, v_n\}. }[/math] - "For the first [math]\displaystyle{ r \le n }[/math] singular values such that si > 0, the left singular vectors are obtained as unit vectors"<ref name="Abdel_SVD"/> by [math]\displaystyle{ \tfrac{1}{s_i}Av_i=u_i. }[/math]
- Select "the [math]\displaystyle{ \ m-r }[/math] left singular vectors corresponding to the zero singular values such that they are unit vectors orthogonal to each other and to the first r left singular vectors"<ref name="Abdel_SVD"/> [math]\displaystyle{ \{u_1, u_2, \dots, u_r\}. }[/math]
Finding the Singular Value Decomposition Using MATLAB Code
Please refer to the following link: http://www.mathworks.com/help/techdoc/ref/svd-singular-value-decomposition.html
Formal definition
"We can now decompose the rectangular matrix A in terms of singular values and vectors as follows"<ref name="Abdel_SVD"/>:
[math]\displaystyle{ A_{m\times n} \begin{bmatrix} v_1 & | & \cdots & | & v_n \end{bmatrix}_{n\times n} = \begin{bmatrix} u_1 & | & \cdots & | & u_n & | I_{n+1} & | & \cdots & | & I_m \end{bmatrix}_{m\times m} \begin{bmatrix} s_1 & 0 & \cdots & 0 \\ 0 & s_2 & \cdots & 0 \\ \vdots & \vdots & \ddots & \vdots \\ 0 & 0 & \cdots & s_n \\ 0 & 0 & 0 & 0 \\ 0 & 0 & 0 & 0 \end{bmatrix}_{m\times n} }[/math]
Where [math]\displaystyle{ \ AV=US }[/math]
Since "the matrices V and U are orthogonal"<ref name="Abdel_SVD"/>, V -1=VT and U -1=UT:
- [math]\displaystyle{ \ A=USV^T }[/math]
"which is the formal definition of the singular value decomposition."<ref name="Abdel_SVD"/>
Relevance to PCA
In order to perform PCA, one needs to do eigenvalue decomposition on the covariance matrix. By transforming the mean for all attributes to zero, the covariance matrix can be simplified to:
[math]\displaystyle{ \ S=XX^T }[/math]
Since the eigenvalue decomposition of ATA gives the same eigenvectors as the singular value decomposition of A, an additional and more consistent method (if a matrix has eigenvectors that are not invertible, its eigenvalue decomposition does not exist) for performing PCA is through the singular value decomposition of X.
The following MATLAB code uses singular value decomposition for performing PCA; 20 principal components, and thus the top 20 maximum variation directions, are selected for reconstructing facial images that have had noise applied to them:
load noisy.mat %first noisy image; each image has a resolution of 20x28 imagesc(reshape(X(:,1),20,28)') %to grayscale colormap gray %singular value decomposition [u s v]=svd(X); %reduced feature space: 20 principal components Xh=u(:,1:20)*s(1:20,1:20)*v(:,1:20)'; figure imagesc(reshape(Xh(:,1),20,28)') colormap gray
Since the reduced feature space image is noiseless, the added noise feature has less variation than the 20 principal components.
References
<references/>
PCA and Introduction to Kernel Function - November 10, 2011
(Continue from the last lecture)
Some notations:
Let [math]\displaystyle{ \displaystyle X }[/math] be a d by n matrix.
Let [math]\displaystyle{ \displaystyle X_j\in\R^d }[/math] be the j th column of [math]\displaystyle{ \displaystyle X, \forall j=1,2,...,n }[/math].
Let [math]\displaystyle{ \displaystyle Q }[/math] be the covariance matrix of [math]\displaystyle{ \displaystyle X }[/math]. So [math]\displaystyle{ \displaystyle Q=(X_j-\bar{X})(X_j-\bar{X})^T }[/math], where [math]\displaystyle{ \bar{X}=\frac{1}{n}\sum_{j=1}^n X_j }[/math]. Strictly speaking, [math]\displaystyle{ \displaystyle Q }[/math] is only the observed covariance matrix from the data collected as the real covariance matrix is unknown.
But we are assuming that we have already centered the data, which means that [math]\displaystyle{ \bar{X}=0 }[/math].
So [math]\displaystyle{ \displaystyle Q_{ij}=(X_i)(X_j)^T=[X X^T]_{ij} }[/math].
Now the principal components of [math]\displaystyle{ \displaystyle X }[/math] are the eigenvectors of [math]\displaystyle{ \displaystyle Q }[/math]. If we do the singular value decomposition, setting [math]\displaystyle{ \ [u\ s\ v] = svd(Q) }[/math], then the columns of [math]\displaystyle{ \ u }[/math] are eigenvectors of [math]\displaystyle{ \displaystyle Q=X X^T }[/math].
Let [math]\displaystyle{ \displaystyle u }[/math] be a [math]\displaystyle{ \displaystyle d\times p }[/math] matrix, where [math]\displaystyle{ \displaystyle p \lt d }[/math], that is composed of the [math]\displaystyle{ \ p }[/math] eigenvectors of [math]\displaystyle{ \displaystyle Q }[/math] corresponding to the largest eigenvalues.
To map the data in a lower dimensional space we project [math]\displaystyle{ \displaystyle X }[/math] onto the [math]\displaystyle{ \ p }[/math] dimensional subspace defined by the columns of [math]\displaystyle{ \displaystyle u }[/math], which are the first [math]\displaystyle{ \ p }[/math] principal components of [math]\displaystyle{ \displaystyle X }[/math].
Let [math]\displaystyle{ \displaystyle Y_{p\times n}={u^T}_{p\times d} X_{d\times n} }[/math]. So [math]\displaystyle{ \displaystyle Y_{p\times n} }[/math] is a lower dimensional approximation of our original data [math]\displaystyle{ \displaystyle X_{d\times n} }[/math]
We can also approximately reconstruct the original data using the dimension-reduced data. However, we will lose some information because when we map those points into lower dimension, we throw away the last [math]\displaystyle{ \ (d - p) }[/math] eigenvectors which contain some of the original information.
[math]\displaystyle{ \displaystyle X'_{d\times n} = {u}_{d\times p} Y_{p\times n} }[/math], where [math]\displaystyle{ \displaystyle X'_{d\times n} }[/math] is an approximate reconstruction of [math]\displaystyle{ \displaystyle X_{d\times n} }[/math].
Example Using Handwritten 2s and 3s
The data X is a 64 by 400 matrix. Every column represents an 8 by 8 image of either a handwritten "2" or "3". The first 200 columns are 2s and the last 200 columns are 3s. First we center the data, and then we find the first 2 eigenvectors of [math]\displaystyle{ \displaystyle XX^T }[/math] using singular value decomposition. Finally we calculate [math]\displaystyle{ \displaystyle Y = u^TX }[/math] and plot the 200 data points in [math]\displaystyle{ \displaystyle Y }[/math]
MATLAB CODE:
MU = repmat(mean(X,2),1,400); % mean(X,2) is the average of all the rows stored in a column vector. % In order to center the data, we extend mean(X,2), which is a 64 by 1 matrix, into a 64 by 400 matrix. Xt = X-MU; % We have modified the data to zero mean data by subtracting the average of each row from every entry in that row. [u s v] = svd(Xt); % Note that size(u) == 64*64, and the columns of u are eigenvectors of VCM. Y = u(:,1:2)'*X; % We project X onto the subspace defined by the first two PCs to get Y. % This transforms the high dimensional points to lower 2 dimensional ones. plot(Y(1,1:200)',Y(2,1:200)','d') hold on plot(Y(1,201:400)',Y(2,201:400)','ro') % We now plot the lower dimensional projection of X. % Essentially we are plotting each point based on the magnitude of Principle Component #1 % and Principle Component #2 in that point. % Note that the first 200 columns represent 2s and are recorded by blue diamonds. % Note that the next 200 columns represent 3s and are recorded with red "o"s.
The result is as follows, we can see clearly there are two classes - the 2s and 3s are generally divided into two sections:
In order to analyze the projection in more detail we can plot the original images on the graph.
image = reshape(X,8,8,400); plotdigits(image,Y,.1,1);
The result can now be seen more clearly from the following picture:
By examining this plot we can infer the approximate "meaning" of the first two principal components in this data. For instance, the points at the top of the plot tend to be slanted to the right, while the ones at the bottom are slanted to the left. So the second principal component quantifies the amount of slant in the number.
Introduction to Kernel Function
PCA is useful when those data points spread in or close to a plane. This means that PCA is powerful when dealing with linear problems. But when data points spread in a manifold space, PCA is hard to implement. But there is a solution to this problem - we can use a method to change the linear algorithm into a nonlinear one. This is called the "Kernel Trick".
An intuitive example
From the picture, we can see the red circles are in the middle of the blue Xs. However, it is hard to separate those two classes by using any linear function (lines in the two dimensional space). But we can use a Kernal function [math]\displaystyle{ \ \phi }[/math] to project the points onto a three dimensional space. Once the blue Xs and red circles have been mapped to a three dimensional space in this way it is easy to separate them using a linear function.
For more details about this trick, please see http://omega.albany.edu:8008/machine-learning-dir/notes-dir/ker1/ker1.pdf
The significance of a Kernel Function, [math]\displaystyle{ \displaystyle \phi }[/math], is that we can implicitly change the data into a high dimension. Let's look at how this is possible:
[math]\displaystyle{ Z_1= \begin{bmatrix} x_1\\ y_1 \end{bmatrix}\xrightarrow{\phi} }[/math] [math]\displaystyle{ \phi(Z_1)= \begin{bmatrix} x_1^2\\ y_1^2\\ \sqrt2x_1y_1 \end{bmatrix} }[/math]
[math]\displaystyle{ Z_2= \begin{bmatrix} x_2\\ y_2 \end{bmatrix}\xrightarrow{\phi} }[/math] [math]\displaystyle{ \phi(Z_2)= \begin{bmatrix} x_2^2\\ y_2^2\\ \sqrt2x_2y_2 \end{bmatrix} }[/math]
The inner product of [math]\displaystyle{ \displaystyle \phi(Z1) }[/math] and [math]\displaystyle{ \displaystyle\phi(Z2) }[/math], which is denoted as [math]\displaystyle{ \displaystyle\phi(Z1)^T\phi(Z2) }[/math], is equal to:
[math]\displaystyle{ \begin{bmatrix} x_1^2&y_1^2&\sqrt2x_1y_1 \end{bmatrix} \begin{bmatrix} x_2^2\\ y_2^2\\ \sqrt2x_2y_2 \end{bmatrix}= }[/math] [math]\displaystyle{ \displaystyle (x_1x_2+y_1y_2)^2=(Z_1^TZ_2)^2=\lt Z_1,Z_2\gt ^2=K(Z_1,Z_2) }[/math].
The most common Kernel functions are as follows:
- Linear: [math]\displaystyle{ \displaystyle K_{ij}=\lt X_i,X_j\gt }[/math]
- Polynomial:[math]\displaystyle{ \displaystyle K_{ij}=(1+\lt X_i,X_j\gt )^p }[/math]
- Gaussian:[math]\displaystyle{ \displaystyle K_{ij}=e^\frac{-{\left\Vert X_i-X_j\right\|}^2}{2\sigma^2} }[/math]
- Exponential:[math]\displaystyle{ \displaystyle K_{ij}=e^\frac{-{\left\Vert X_i-X_j\right\|}}{2\sigma^2} }[/math]
Note: [math]\displaystyle{ {\left\Vert X_i-X_j\right\|} }[/math] denotes the distance between [math]\displaystyle{ \displaystyle X_i }[/math] and [math]\displaystyle{ \displaystyle X_j }[/math].
Disadvantages of the Kernel Trick
Since the kernel trick brings a lower dimensional problem to a higher dimensional space, it is affected by the curse of dimensionality. The curse of dimensionality states that it takes exponentially more points to estimate a solution in higher dimensions. ex) if [math]\displaystyle{ \R^1 }[/math] needs 10 points, then [math]\displaystyle{ \R^2 }[/math] needs 100, [math]\displaystyle{ \R^3 }[/math] needs 1000 etc.
Kernel PCA - November 15, 2011
PCA doesn't work well when the directions of variation in our data are nonlinear. Especially in the case where the dataset has very high dimensions, the dataset lies near or on a nonlinear manifold which prevents PCA from determining principal components correctly. To deal with this problem, we apply kernels to PCA. By transforming the original data with a nonlinear mapping, we can obtain much better principal components.
First we look at the algorithm for PCA and see how we can kernelize PCA:
PCA
Find eigenvectors of [math]\displaystyle{ \ XX^T }[/math], call it [math]\displaystyle{ \ U }[/math]
to map data points to a lower dimensional space:
[math]\displaystyle{ \ Y = U^{T}X }[/math]
to reconstruct points:
[math]\displaystyle{ \ \hat{X} = UY }[/math]
to map a new point:
[math]\displaystyle{ \ y = U^{T}x }[/math]
to reconstruct point:
[math]\displaystyle{ \ \hat{x} = Uy }[/math]
Dual PCA
Consider the singular value decomposition of n-by-m matrix X:
[math]\displaystyle{ \begin{align} \left[ U\ \Sigma\ V \right] & = svd(X) \\ X & = U\Sigma{V^T} \end{align} }[/math]
Where:
- The columns of U are the eigenvectors of [math]\displaystyle{ XX^T }[/math] corresponding to the eigenvalues in decreasing order.
- The columns of V are the eigenvectors of [math]\displaystyle{ X^T{X} }[/math].
- The diagonal matrix [math]\displaystyle{ \Sigma }[/math] contains the square roots of the eigenvalues of [math]\displaystyle{ XX^T }[/math] in decreasing order.
Now we want to kernelize this classical version of PCA. We would like to express everything based on V which can be kernelized. The reason why we want to do this is because if n >> m (i.e. the feature space is much larger than the number of sample points) then the original PCA algorithm would be impractical. We want an algorithm that depends less on n. This is called Dual PCA.
U in terms of V:
[math]\displaystyle{ \begin{align} X &= U\Sigma V^T \\ XV &= U\Sigma V^T V = U\Sigma \\ XV\Sigma^{-1} &= U\Sigma\Sigma^{-1} = U \\ U &= XV\Sigma^{-1} \end{align} }[/math]
Y in terms of V:
[math]\displaystyle{ \begin{align} X&=U \Sigma V^T \\ U^TX &= U^TU\Sigma V^T \\ U^TX &= \Sigma V^T \\ Y&=\Sigma V^T \\ \end{align} }[/math]
Reconstructed points in terms of V:
[math]\displaystyle{ \begin{align} \hat{X}&=UY \\ &=XV\Sigma^{-1}\Sigma{V^T} \\ &= XVV^T \\ &= X \end{align} }[/math]
The value of y, a single point in the sample reduced to low-dimensional space, in terms of V:
[math]\displaystyle{ \begin{align} y &= U^Tx \\ & = (XV\Sigma^{-1})^Tx \\ & = (\Sigma^{-1})^T{V^T}{x^T}x \\ & = \Sigma^{-1}{V^T}{x^T}x \end{align} }[/math]
A single reconstructed point from the sample in terms of V:
[math]\displaystyle{ \begin{align} \hat{x} &= Uy \\ & = UU^Tx \\ & =XV\Sigma^{-1}\Sigma^{-1}V^T{x^T}x \\ &= XV\Sigma^{-2}V^T{x^T}x \end{align} }[/math]
Kernel PCA
The nonlinear mapping [math]\displaystyle{ \displaystyle\phi }[/math] allows for very high dimensional spaces and is never calculated explicitly.
- [math]\displaystyle{ k(\mathbf{x},\mathbf{x}) = \phi^T(\mathbf{x})\phi(\mathbf{x}) }[/math]
[math]\displaystyle{ X_{d\times n} }[/math], [math]\displaystyle{ X^TX_{n\times n} }[/math], and [math]\displaystyle{ K(X,X)_{n\times n} }[/math] could have many different kernels.
Generally, we want to replace [math]\displaystyle{ X^TX }[/math] with a kernel. The idea in Kernel PCA is that instead of finding the eigenvectors of [math]\displaystyle{ X^TX }[/math], we can find the eigenvectors of a kernel.
Example.[math]\displaystyle{ \displaystyle K(x_1,x_2)=e^\frac{-(X_1-X_2)^2}{\sigma} }[/math]
Find the eigenvectors of the matrix [math]\displaystyle{ K=\left(\begin{matrix}0&0\dots&\dots\\ \vdots&\ddots\\\vdots\\\end{matrix}\right) }[/math]
However, since the [math]\displaystyle{ \displaystyle\phi }[/math] is never calculated explicitly, kernel PCA cannot reconstruct points using the equation [math]\displaystyle{ \hat{X} = XVV^T }[/math] where [math]\displaystyle{ X=\phi }[/math].
In kernel PCA, we replace [math]\displaystyle{ k }[/math] for [math]\displaystyle{ \phi(X)^T \phi(X) }[/math]. This is correct if [math]\displaystyle{ \phi(X) }[/math] has a mean of zero.
We need to find a way to centralize [math]\displaystyle{ \phi(X) }[/math].
Centralizing Kernel PCA
Recall in regular PCA, our variance-covariance matrix [math]\displaystyle{ \Sigma }[/math] was defined as [math]\displaystyle{ \Sigma = (X-\mu)(X-\mu)^T }[/math].
With kernel PCA, we will never explicitly define [math]\displaystyle{ \phi(x) }[/math] so we need a method to calculate its mean without calculating the actual transformation of [math]\displaystyle{ \phi(x) }[/math] itself.
We define:
- [math]\displaystyle{ \tilde{\phi}(x) = \phi(x) - E_x[\phi(x)] }[/math]
And thus, our kernel function becomes:
- [math]\displaystyle{ \tilde{k}(\mathbf{x},\mathbf{y}) = \tilde{\phi}(\mathbf{x})^T\tilde{\phi}(\mathbf{y}) }[/math]
- [math]\displaystyle{ \! = (\phi(x) - E_x[\phi(x)])^T(\phi(y) - E_y[\phi(y)]) }[/math]
- [math]\displaystyle{ \! = \phi(x)^T\phi(y) - \phi(y)E_x[\phi(x)^T] - \phi(x)^TE_y[\phi(y)] + E_x[\phi(x)^T]E_y[\phi(y)] }[/math]
- [math]\displaystyle{ \! = k(\mathbf{x},\mathbf{y}) - E_x[k(\mathbf{x},\mathbf{y})] - E_y[k(\mathbf{x},\mathbf{y})] + E_x[E_y[k(\mathbf{x},\mathbf{y})]] }[/math]
In practice, we would do the following:
1. Start with our matrix of data: [math]\displaystyle{ X_{d\times n} }[/math]
2. Choose a kernel function [math]\displaystyle{ k }[/math]
3. Compute [math]\displaystyle{ K }[/math], an nxn matrix: [math]\displaystyle{ K=\left(\begin{matrix}k(x_1,x_1)&k(x_1,x_2)\dots&k(x_1,x_n)\\ \vdots&\ddots&\vdots\\k(x_n,x_1)&\dots&k(x_n,x_n)\\\end{matrix}\right) }[/math]
4. Find [math]\displaystyle{ \tilde{K} = K - \frac{1}{n}\sum_{i=1}^{n}K(:,i) - \frac{1}{n}\sum_{j=1}^{n}K(j,:) + \frac{1}{n^2}\sum_{i=1}^{n}\sum_{j=1}^{n}K(i,j) }[/math]
Note: On the right hand side, the second item is the average of the columns of [math]\displaystyle{ K }[/math], the third item is the average of the rows of [math]\displaystyle{ K }[/math], and the last item is the average of all the entries of [math]\displaystyle{ K }[/math]. As a result, they are vectors of dimension [math]\displaystyle{ 1\times n }[/math], [math]\displaystyle{ n\times 1 }[/math], and [math]\displaystyle{ 1\times 1 }[/math] (a scalar) respectively. To do the actual arithmetic, all matrix dimensions must match. In MATLAB, this is accomplished through the repmat() function.
5. Find the eigenvectors of [math]\displaystyle{ \tilde{K} }[/math]. Take the first few and combine them in a matrix [math]\displaystyle{ V }[/math], and use that as the [math]\displaystyle{ V }[/math] in the dual PCA mapping and reconstruction steps shown above.
Multidimensional Scaling (MDS)
Most of the common linear and polynomial kernels do not give very good results in practice. For real data, there is an alternate approach that can yield much better results but, as we will see later, is equivalent to a kernel PCA process.
Multidimensional Scaling (MDS) is an algorithm, much like PCA, that maps the original high dimensional space to a lower dimensional space.
Introduction to MDS
The main purpose of MDS is to try to map high dimensional data to a lower dimensional space while preserving the pairwise distances between points. ie: MDS addresses the problem of constructing a configuration of [math]\displaystyle{ \ n }[/math] points in Euclidean space by using information about the distance between the [math]\displaystyle{ \ n }[/math] patterns.
It may not be possible to preserve the exact distances between points, so in this case we need to find a representation that is as close as possible.
Definition: A [math]\displaystyle{ \ n\times n }[/math] matrix [math]\displaystyle{ \ D }[/math] is called the distance matrix if:
[math]\displaystyle{ \ D }[/math] is symmetric and its entries [math]\displaystyle{ \ d_{ij} }[/math] have the properties:
[math]\displaystyle{ \ d_{ii} = 0 }[/math] and [math]\displaystyle{ \ d_{ij} \gt 0 }[/math] [math]\displaystyle{ \forall i\neq j }[/math]
Given a distance matrix [math]\displaystyle{ \ D^{(X)} }[/math], MDS attempts to find [math]\displaystyle{ \ n }[/math] data points [math]\displaystyle{ \ y_1,...,y_n }[/math] in [math]\displaystyle{ \ d }[/math] dimensions, such that if [math]\displaystyle{ \ d_{ij}^{(Y)} }[/math] denotes the Euclidean distance between [math]\displaystyle{ \ y_i, y_j }[/math] then [math]\displaystyle{ \ D^{(Y)} }[/math] is similar to [math]\displaystyle{ \ D^{(X)} }[/math].
Metric MDS
One of the possible methods to preserve the pairwise distance is to use Metric MDS, which attempts to minimize: [math]\displaystyle{ \min_Y \sum_{i=1}^{n}\sum_{j=1}^{n}(d_{ij}^{(X)} - d_{ij}^{(Y)})^2 }[/math]
where [math]\displaystyle{ d_{ij}^{(X)} = ||x_i-x_j||^2 }[/math] and [math]\displaystyle{ d_{ij}^{(Y)} = ||y_i-y_j||^2 }[/math]
Continued with MDS, Isomap and Classification - November 22, 2011
The distance matrix can be converted to a kernel matrix of inner products [math]\displaystyle{ \ X^TX }[/math] by [math]\displaystyle{ \ X^{T}X=-\frac{1}{2}HD^{(X)}H }[/math],
where [math]\displaystyle{ H=I-\frac{1}{n}ee^{T} }[/math] and [math]\displaystyle{ \ e }[/math] is a column vector of all 1.
[math]\displaystyle{
e=\left[\begin{array}{c}
1\\
\vdots\\
1
\end{array}\right]_{(n\times1)}
}[/math]
[math]\displaystyle{ ee^{T}=\left[\begin{array}{ccc}
1 & \cdots & 1\\
\vdots & \ddots & \vdots\\
1 & \cdots & 1
\end{array}\right]_{(n\times n)}
}[/math]
[math]\displaystyle{
H=\left[\begin{array}{ccc}
1 & \cdots & 0\\
\vdots & \ddots & \vdots\\
0 & \cdots & 1
\end{array}\right]-\frac{1}{n}\left[\begin{array}{ccc}
1 & \cdots & 1\\
\vdots & \ddots & \vdots\\
1 & \cdots & 1
\end{array}\right]
}[/math]
Now [math]\displaystyle{ \min_Y \sum_{i=1}^{n}\sum_{j=1}^{n}(d_{ij}^{(X)} - d_{ij}^{(Y)})^2 }[/math] can be reduced to [math]\displaystyle{ \min_Y \sum_{i=1}^{n}\sum_{j=1}^{n}(x_{i}^{T}x_{j}-y_{i}^{T}y_{j})^{2} }[/math] [math]\displaystyle{ \Leftrightarrow }[/math][math]\displaystyle{ \min_Y \ Tr(X^{T}X-Y^{T}Y)^{2} }[/math].
So far we have rewritten the objective function, but we must remember that our definition of [math]\displaystyle{ \ D }[/math] introduces constraints on its components. We must ensure that these constraints are respected in the way we have expressed the minimization problem with traces. Luckily, we can capture these constraints with a single requirement thanks to the following theorem:
Theorem: Let [math]\displaystyle{ \ D }[/math] be a distance matrix and [math]\displaystyle{ \ K=X^{T}X=-\frac{1}{2}HD^{(X)}H }[/math]. Then [math]\displaystyle{ \ D }[/math] is Euclidean if and only if [math]\displaystyle{ \ K }[/math] is a positive semi-definite matrix.
Therefore, to complete the rewriting of our original minimization problem from norms to traces, it suffices to impose that [math]\displaystyle{ \ K }[/math] be [math]\displaystyle{ \ p.s.d. }[/math], as this guarantees that [math]\displaystyle{ \ D }[/math] is a distance matrix and the components of [math]\displaystyle{ \ X^{T}X }[/math] and [math]\displaystyle{ \ Y^{T}Y }[/math] satisfy the original constraints.
Proceeding with singular value decomposition, [math]\displaystyle{ \ X^{T}X }[/math] and [math]\displaystyle{ \ Y^{T}Y }[/math] can be decomposed as:
[math]\displaystyle{ \ X^{T}X=V\Lambda V^{T} }[/math]
[math]\displaystyle{ Y^{T}Y=Q\hat{\Lambda} Q^{T} }[/math]
Since [math]\displaystyle{ \ Y^{T}Y }[/math] is [math]\displaystyle{ \ p.s.d. }[/math] , [math]\displaystyle{ \hat{\Lambda} }[/math] has no negative value and therefore: [math]\displaystyle{ Y=\hat{\Lambda}^{\frac{1}{2}}Q^{T} }[/math].
The above definitions help rewrite the cost function as:
[math]\displaystyle{ \min_{Q,\hat{\Lambda}}Tr(V\Lambda V^{T}-Q\hat{\Lambda}Q^{T})^{2} }[/math]
Multiply [math]\displaystyle{ \ V^{T} }[/math] on the left and [math]\displaystyle{ \ V }[/math] on the right, we will get
[math]\displaystyle{ \min_{Q,\hat{\Lambda}}Tr(\Lambda-V^{T}Q\hat{\Lambda} Q^{T}V)^{2} }[/math]
Then let [math]\displaystyle{ \ G=V^{T}Q }[/math]
We can rewrite the target function as:
[math]\displaystyle{ \min_{Q,\hat{\Lambda}}Tr(\Lambda-G\hat{\Lambda}G^{T})^{2} }[/math]
[math]\displaystyle{ = }[/math][math]\displaystyle{ \min_{G,\hat{\Lambda}}Tr(\Lambda^{2}+G{\Lambda}G^{T}G\hat{\Lambda}G^{T}-2\Lambda G\hat{\Lambda}G) }[/math]
For a fixed [math]\displaystyle{ \hat{\Lambda} }[/math] we can minimize for G. The result is that [math]\displaystyle{ \ G=I }[/math]. Then we can simplify the target function:
[math]\displaystyle{ \min_{\hat{\Lambda}}Tr(\Lambda^{2}+\hat{\Lambda}^{2}-2\Lambda\hat{\Lambda})
}[/math] [math]\displaystyle{ = }[/math] [math]\displaystyle{ \min_{\hat{\Lambda}}Tr(\Lambda-\hat{\Lambda})^{2} }[/math]
Obviously, [math]\displaystyle{ \hat{\Lambda}=\Lambda }[/math].
If [math]\displaystyle{ \ X }[/math] is [math]\displaystyle{ d\times n }[/math] matrix, [math]\displaystyle{ \ (d\lt n) }[/math], the rank of [math]\displaystyle{ \ X }[/math] is no greater than [math]\displaystyle{ \ d }[/math]. Then the rank of [math]\displaystyle{ X^{T}X_{(n\times n)} }[/math] is no greater than [math]\displaystyle{ \ d }[/math] since [math]\displaystyle{ \ rank(X^{T}X) = rank(X) }[/math]. The dimension of [math]\displaystyle{ \ Y }[/math] is smaller than [math]\displaystyle{ \ X }[/math], so therefore the rank of [math]\displaystyle{ \ Y^{T}Y }[/math] is smaller than [math]\displaystyle{ \ d }[/math].
Since we want to do dimensionality reduction and make [math]\displaystyle{ \ \Lambda }[/math] and [math]\displaystyle{ \hat{\Lambda} }[/math] as similar as possible, we can let [math]\displaystyle{ \hat{\Lambda} }[/math] be the top [math]\displaystyle{ \ d }[/math] diagonal elements of [math]\displaystyle{ \ \Lambda }[/math].
We also have [math]\displaystyle{ \ G = V^{T}Q }[/math], obviously [math]\displaystyle{ \ Q=V }[/math] since [math]\displaystyle{ \ G = I }[/math].
The solution is:
[math]\displaystyle{ Y=\Lambda^{\frac{1}{2}}V^{T} }[/math]
where [math]\displaystyle{ \ V }[/math] is the eigenvector of [math]\displaystyle{ \ X^{T}X }[/math] corresponding to the top [math]\displaystyle{ \ d }[/math]
eigenvalues, and [math]\displaystyle{ \Lambda }[/math] is the top [math]\displaystyle{ \ d }[/math] eigenvalues of [math]\displaystyle{ \ X^{T}X }[/math].
Compare this with dual PCA.
In dual PCA, the result is
[math]\displaystyle{ \ Y=\Sigma V^{T} }[/math].
Clearly, the result of dual PCA is the same with MDS. Actually, one property of PCA is to preserve the pairwise distances between data points in both high dimensional and low dimensional space.
Now as an appendix, we provide a short proof to the first equation [math]\displaystyle{ \ X^{T}X=-\frac{1}{2}HD^{(X)}H }[/math].
Let [math]\displaystyle{ \ S }[/math] be a vector where [math]\displaystyle{ S_{i}=X_i^TX_i,D=Se^T+eS^T-2X^TX }[/math].
First notice [math]\displaystyle{ \ Xe=0 }[/math] as data are centered.
[math]\displaystyle{ HD^{(X)}H=(I-\frac{1}{n}ee^{T})(Se^T+eS^T-2X^TX)(I-\frac{1}{n}ee^{T}) }[/math]
[math]\displaystyle{ =-2X^TX+\frac{1}{n}ee^{T}2X^TX+\frac{1}{n}2X^TXee^{T}-\frac{1}{n^2}ee^{T}2X^TXee^{T} }[/math]
[math]\displaystyle{ \ =-2X^TX }[/math],
as all other terms contains [math]\displaystyle{ \ Xe }[/math] or [math]\displaystyle{ \ e^TX^T }[/math].
Hence, [math]\displaystyle{ \ X^{T}X=-\frac{1}{2}HD^{(X)}H }[/math].
Isomap (As per Handout - Section 1.6)
The Isomap algorithm is a nonlinear generalization of classical MDS with the main idea being that MDS is perfomed on the geodesic space of the non-linear data manifold as opposed to being performed on the input space. Isomap applies geodesic distances in the distance matrix for MDS rather than the straight line distances between the 2 points in order to find the low-dimensional mapping that preserves the pairwise distances. This geodesic distance is approximated by building a k-neighbourhood graph of all the points on the manifold and finding the shortest path to a given point. This gives a much better distance measurement in a manifold such as the 'Swiss Roll' manifold than using Euclidean distances.
Note: Geodesic distance is the shortest path along the curved surface of the manifold measured as if the surface was flat
Similarly to LLE, the Isomap algorithm proceeds in three steps:
1. Finding the neighbours of each data points in high-dimensional data space
2. Compute the geodesic pairwise distance between all points
3. Integrate the data via MDS in order to preserve the distances
Process:
1. Identify the k nearest neighbours or choose points from a fixed radius
2. Neighbour relations are represented by a graph G conneted with edges of weights [math]\displaystyle{ \ d_{X}(i,j) }[/math]
3. The geodesic distances, [math]\displaystyle{ \ d_{M}(i,j) }[/math] between all pair points on the manifold, M, are then estimated
Note: Isomap approximates [math]\displaystyle{ \ d_{M}(i,j) }[/math] as the shortest path distance [math]\displaystyle{ \ d_{G}(i,j) }[/math] in Graph G. The k in the algorithm must be chosen carefully, too small a k and the graph will not be connected, but to large a k, and the algorithm will be closer to the euclidean distance instead.
4. Isomap applies classical MDS to [math]\displaystyle{ \ D^{(G)} }[/math] to generate an embedding of the data in d-dimensional Euclidean space Y
Classification
Classification is a technique in pattern recognition. It can yield very complex decision boundaries as they are very suitable for ordered data, categorical data or a mixture of the two types. A decision or classification represents a multi-stage decision process where a binary decision is made at each stage.
E.g: A hand-written object can be scanned and recognized by the classification technique. The model realized the class it belongs to and pairs it with the corresponding object in its library.
Mathematically, each object have a set of features [math]\displaystyle{ \ X }[/math] and a corresponding label [math]\displaystyle{ \ Y }[/math] which is the class it belongs to:
[math]\displaystyle{ \ \{(x_1,y_1),(x_2,y_2),...,(x_n,y_n)\} }[/math]
Since the training set is labelled with the correct answers, classification is called a "supervised learning" method.
In contrast, the Clustering technique is used to explore a data set whereby the main objective is to separate the sample into groups or to provide an understanding about the underlying structure or nature of the data. Clustering is an "unsupervised classification" method, because we do not know the groups that are in the data or any group characteristics of any unit observation. There are no labels to classify the data points, all we have is the feature set [math]\displaystyle{ \ X }[/math]:
[math]\displaystyle{ \ \{(x_1),(x_2),...,(x_n)\} }[/math]
Classification - November 24, 2011
Classification
Classification is predicting a discrete random variable Y (the label) from another random variable X. It is analogous to regression, but the difference between them is that regression uses continuous values, while classification uses discrete values (labels).
Consider iid data [math]\displaystyle{ \displaystyle (X_1,Y_1),(X_2,Y_2),...,(X_n,Y_n) }[/math] where
[math]\displaystyle{ X_i = (X_{i1},...,X_{id}) \in \mathbb{R}^{d} }[/math], representing an object,
[math]\displaystyle{ \ Y_i \in Y }[/math] is the label of the i-th object, and [math]\displaystyle{ \ Y }[/math] is some finite set.
We wish to determine a function [math]\displaystyle{ \ h : \mathbb{R}^{d} \rightarrow Y }[/math] that can predict the value of [math]\displaystyle{ \ Y_i }[/math] given the value [math]\displaystyle{ \ X_i }[/math]. When we observe a new [math]\displaystyle{ \displaystyle X }[/math], predict [math]\displaystyle{ \displaystyle Y }[/math] to be [math]\displaystyle{ \displaystyle h(X) }[/math].(We use h(x)) for the following discussion).
The difference between classification and clustering is that clustering do not have [math]\displaystyle{ \ Y_i }[/math] and it only puts [math]\displaystyle{ \ X_i }[/math] into different classes.
Examples
Object: An image of a pepper'
The features are defined to be colour, length, diameter, and weight.
File:pepper.jpg
Features(X): (Red,6,2,3.5)
Labels(Y): Red Pepper
The objective of classification is to use the classification function for unseen data - data for which we do not know the classification. Given the features, we want to classify the object, or in other words find the label. This is the typical classification problem.
Real Life Examples: Face Recognition - Pictures are just a points in high dimensional space; we can represent them in vectors. Sound waves can also be classified in a similar manner using Fourier Transforms. Classification is also used to find drugs which cure disease. Molecules are classified as either a good fit into the cavity of a protein, or not.
In machine learning, classification is also known as supervised learning. Often we separate the data set into two parts. One is called a training set and the other, a testing set. We use the training set to establish a classifier rule and use the testing set to test its effectiveness.
Error Rate
Definition: The true error rate of a classifier h is [math]\displaystyle{ L(h) = P(h(X) \neq Y) }[/math] and the empirical error rate or training error rate is:
[math]\displaystyle{ \hat{L}_h = \frac{1}{n} \sum_{i=1}^{n}I(h(X_i)\neq Y_i) }[/math]
Where [math]\displaystyle{ \!I }[/math] is the indicator function. That is, [math]\displaystyle{ \!I( ) = 1 }[/math] if the statement inside the bracket is true, and [math]\displaystyle{ \!I( ) = 0 }[/math] otherwise.
The empirical error rate is the the proportion of points that have not been classified correctly. It can be shown that this estimation always underestimates the true error rate, although we do not cover it in this course.
A way to get a better estimate of the error is to construct the classifier with half of the given data and use the other half to calculate the error rate.
Bayesians vs Frequentists
Bayesians view probability as the measure of confidence that a person holds in a proposition given certain information. They state a "prior probability" exist, which represents the possibility of a event's occurrence given no information. As we add new information regarding this event, our belief adjusts according to the new information, which gives us a posterior probability. Frequentists interpret probability as a "propensity" of some event.
Bayes Classifier
Consider the special case where [math]\displaystyle{ y \in {0,1} }[/math], then
[math]\displaystyle{ r(x) = P(Y=1|X=x) = \frac{P(X=x|Y=1)P(Y=1)}{P(X=x)} = \frac{P(X=x|Y=1)P(Y=1)}{P(X=x|Y=1)P(Y=1)+P(X=x|Y=0)P(Y=0)} }[/math]
Definition: The Bayes classification rule h* is:
[math]\displaystyle{ h^*(x)=\left\{\begin{matrix}1 & \mathrm{if}\ \hat{r}(x) \gt 0.5 \\ 0 & otherwise\end{matrix}\right. }[/math]
The set [math]\displaystyle{ \displaystyle D(h) = \{x: P(Y=1|X=x)=P(Y=0|X=x)\} }[/math] is called the decision boundary.
[math]\displaystyle{ h^*(x)=\left\{\begin{matrix}1 & \mathrm{if}\ P(Y=1|X=x) \gt P(Y=0|X=x) \\ 0 & otherwise\end{matrix}\right. }[/math]
Theorem: The Bayes rule is optimal, that is, if [math]\displaystyle{ \ h }[/math] is any other classification rule, then [math]\displaystyle{ L(h^*) \leq L(h) }[/math]
So if Bayes rule is optimal, why do we need any other method? Well, we don't always know the distributions of the data. The Bayes rule depends on unknown quantities, so we need to use the data to find some approximation to the Bayes rule.
Three Main Approaches
1. Empirical Risk Minimization: Choose a set of classifiers H and find a [math]\displaystyle{ h^* \in H }[/math] that minimizes the expected value of some loss function [math]\displaystyle{ \ L(h) }[/math]. The distribution of (X,Y) is not known, therefore [math]\displaystyle{ \ E[L(h)] }[/math] must be estimated using empirical data. For more conceptual knowledge on how this approach works, please refer to http://en.wikipedia.org/wiki/Empirical_risk_minimization
2. Regression: Find an estimate [math]\displaystyle{ \hat{r}(x) }[/math] of the function [math]\displaystyle{ \displaystyle r }[/math] and define:
[math]\displaystyle{ h^*(x)=\left\{\begin{matrix}1 & \mathrm{if}\ \hat{r}(x) \gt 0.5 \\ 0 & otherwise\end{matrix}\right. }[/math]
3. Density Estimation: Estimate [math]\displaystyle{ \displaystyle P(X=x|Y=0) }[/math] from the [math]\displaystyle{ \displaystyle X_i's }[/math] for which [math]\displaystyle{ \displaystyle Y_i=0 }[/math], and estimate [math]\displaystyle{ \displaystyle P(X=x|Y=1) }[/math] from the [math]\displaystyle{ \displaystyle X_i's }[/math] for which [math]\displaystyle{ \displaystyle Y_i=1 }[/math] and let:
[math]\displaystyle{ \displaystyle P(Y=1)=\frac{1}{n} \sum_{i=1}^{n}Y_i }[/math]
Define
[math]\displaystyle{ \hat{r}(x)=\hat{P}(Y=1|X=x) }[/math]
and define
[math]\displaystyle{ h(x)=\left\{\begin{matrix}1 & \mathrm{if}\ \hat{r}(x) \gt 0.5 \\ 0 & otherwise\end{matrix}\right. }[/math]
So we have
[math]\displaystyle{ P(Y=1|X=x) = \frac{P(X=x|Y=1)P(Y=1)}{P(X)} }[/math]
where
[math]\displaystyle{ \displaystyle P(X) = P(X=x|Y=1)P(Y=1) + P(X=x|Y=0)P(Y=0) }[/math]
Multi-class Classification
We want to generalize to the case that Y takes on more than two values
Theorem: Suppose that [math]\displaystyle{ Y \in y = \{1,...,k\} }[/math], the optimal rule is
[math]\displaystyle{ \displaystyle h^*(x) = {\operatorname{arg\,max}}_k\{P(Y=k|X=x)\} }[/math]
where
[math]\displaystyle{ P(Y=k|X=x)=\frac{f_k(x)\pi_k}{\sum_{k}f_k(x)\pi_k} }[/math]
with
[math]\displaystyle{ \displaystyle f_k(x) = P(X=x|Y=k) }[/math]
[math]\displaystyle{ \displaystyle \pi_k = P(Y=k) }[/math]
A direct interpretation of this method called "naive Bayes classifier", is that the classifier chooses an y value which maximizes the probability of getting the current x value. In other words, the value y here can be viewed as a parameter as in maximum likelihood estimation, where the optimal value is [math]\displaystyle{ \displaystyle y^* = {\operatorname{arg\,max}}\{f(y|x)\} }[/math].
LDA, QDA and FDA - November 29, 2011
Linear Discriminant Analysis (LDA) and Quadratic Discriminant Analysis (QDA)
The simplest approach to classification is to use the third approach listed above and assume a parametric model for densities.
Applying Bayes rule:
[math]\displaystyle{ P(Y=k|X=x) = \frac{P(X=x|Y=k)P(Y=k)}{P(X=x)} = \frac{P(X=x|Y=k)P(Y=k)}{\sum_{i}P(X=x|Y=i)P(Y=i)} }[/math]
We notice that the denominator value is always the same independent of k; thus will be cancelled out. Hence we will only need to evaluate the numerator value.
Define class conditional distribution, prior distribution and posterior distribution as follows:
[math]\displaystyle{ \ f_k(x)=P(X=x|Y=k),{\mathbf\pi_k}=P(Y=k), P(Y=k|X=x) }[/math]
LDA (Linear Discriminant Analysis)
LDA is a classifier which makes the 2 assumptions:
- The class conditional distribution is Gaussian
- [math]\displaystyle{ \ f_k(x)= \frac{1}{(2\pi)^{d/2}|\Sigma_k|^{1/2}} \exp\left(-\frac{1}{2}({\mathbf x}-{\mathbf\mu_k})^T{\mathbf\Sigma_k}^{-1}({\mathbf x}-{\mathbf\mu_k}) \right),x\in \mathbb{R}^d }[/math]
where [math]\displaystyle{ |\mathbf\Sigma_k| }[/math] is the determinant of [math]\displaystyle{ \mathbf\Sigma_k }[/math]
- The two classes share the same covariance matrix
- [math]\displaystyle{ {\mathbf\Sigma}_0={\mathbf\Sigma}_1={\mathbf\Sigma} }[/math]
Thus the class conditional distribution for [math]\displaystyle{ k }[/math] in LDA is as follows:
[math]\displaystyle{ \ f_k(x)= \frac{1}{(2\pi)^{d/2}|\Sigma|^{1/2}}
\exp\left(-\frac{1}{2}({\mathbf x}-{\mathbf\mu_k})^T{\mathbf\Sigma}^{-1}({\mathbf x}-{\mathbf\mu_k})
\right),x\in \mathbb{R}^d }[/math]
To demonstrate, suppose we have a two class scenario, i.e [math]\displaystyle{ y \in \{0,1 \} }[/math], and we want to compute the decision boundary for this LDA. Recall that the decision boundary is a set of points such that:
[math]\displaystyle{ \ P(Y=0|X=x)=P(Y=1|X=x) }[/math]
where the probability is given by:
[math]\displaystyle{ P(Y=0|X=x) = \frac{f_0(x){\mathbf\pi_0}}{\sum_{k=0}^{1}f_k(x){\mathbf\pi_k}},P(Y=1|X=x) = \frac{f_1(x){\mathbf\pi_1}}{\sum_{k=0}^{1}f_k(x){\mathbf\pi_k}} }[/math]
Hence we want to find [math]\displaystyle{ {\mathbf x} }[/math] such that
[math]\displaystyle{ \ P(Y=0|X=x)=P(Y=1|X=x) }[/math]
[math]\displaystyle{ \frac{f_0(x){\mathbf\pi_0}}{\sum_{k=0}^{1}f_k(x){\mathbf\pi_k}} = \frac{f_1(x){\mathbf\pi_1}}{\sum_{k=0}^{1}f_k(x){\mathbf\pi_k}} }[/math]
[math]\displaystyle{ f_0(x){\mathbf\pi_0}=f_1(x){\mathbf\pi_1} }[/math]
Upon expanding [math]\displaystyle{ \ f_k(x) }[/math], we get
[math]\displaystyle{
\frac{1}{(2\pi)^{d/2}|\Sigma|^{1/2}}
\exp\left(-\frac{1}{2}({\mathbf x}-{\mathbf\mu_0})^T{\mathbf\Sigma}^{-1}({\mathbf x}-{\mathbf\mu_0})
\right) * \pi_0
}[/math]
[math]\displaystyle{
=\frac{1}{(2\pi)^{d/2}|\Sigma|^{1/2}}
\exp\left(-\frac{1}{2}({\mathbf x}-{\mathbf\mu_1})^T{\mathbf\Sigma}^{-1}({\mathbf x}-{\mathbf\mu_1})
\right) * \pi_1
}[/math]
Rearranging the equation and taking logarithm on both sides we get:
[math]\displaystyle{
-\frac{1}{2}({\mathbf x}-{\mathbf\mu_0})^T{\mathbf\Sigma}^{-1}({\mathbf x}-{\mathbf\mu_0})+\log({\mathbf\pi_0})
=-\frac{1}{2}({\mathbf x}-{\mathbf\mu_1})^T{\mathbf\Sigma}^{-1}({\mathbf x}-{\mathbf\mu_1})+\log({\mathbf\pi_1})
}[/math]
That is, after expanding the quadratic form of our vector values,
[math]\displaystyle{
\frac{1}{2}
\left
[-{\mathbf x}^T{\mathbf\Sigma}^{-1}{\mathbf x}
-{\mathbf\mu_1}^T{\mathbf\Sigma}^{-1}{\mathbf\mu_1}
+2{\mathbf x}^T{\mathbf\Sigma}^{-1}{\mathbf\mu_1}
+{\mathbf x}^T{\mathbf\Sigma}^{-1}{\mathbf x}
+{\mathbf\mu_0}^T{\mathbf\Sigma}^{-1}{\mathbf\mu_0}
-2{\mathbf x}^T{\mathbf\Sigma}^{-1}{\mathbf\mu_0}
\right]
+\log(\frac{\mathbf\pi_1}{\mathbf\pi_0})
=0
}[/math]
Cancelling out the terms and notice that [math]\displaystyle{ {\mathbf\mu_k}^T{\mathbf\Sigma}^{-1}{\mathbf\mu_k} }[/math] is constant and [math]\displaystyle{ {\mathbf x}^T{\mathbf\Sigma}^{-1}{\mathbf\mu_k} }[/math] a linear expression of [math]\displaystyle{ \ x }[/math], the final equation is simply linear system of equations:
[math]\displaystyle{ {\mathbf x}^T{\mathbf\Sigma}^{-1}({\mathbf\mu_1}-{\mathbf\mu_0})
+\frac{1}{2}\left(-{\mathbf\mu_1}^T{\mathbf\Sigma}^{-1}{\mathbf\mu_1}
+{\mathbf\mu_0}^T{\mathbf\Sigma}^{-1}{\mathbf\mu_0}\right)
+\log(\frac{\mathbf\pi_1}{\mathbf\pi_0})
=0
}[/math]
QDA (Quadratic Discriminant Analysis)
If we relax the assumption in LDA that the covariances are identical between classes, the decision boundary becomes quadratic and we get QDA. Mathematically we mean that [math]\displaystyle{ {\mathbf\Sigma}_0\ne{\mathbf\Sigma}_1 }[/math]. In fact, we may also drop the assumption of a bi-class problem. QDA is therefore a more generalized version of LDA.
The general form of the boundary for a K-class problem is
[math]\displaystyle{ \delta_k (x) = -\frac{1}{2}ln|{\mathbf\Sigma}_k| -\frac{1}{2}(x-{\mathbf\mu_k})^T{\mathbf\Sigma_k}^{-1} (x-{\mathbf\mu_k}) + ln({\mathbf\pi_k}) }[/math]
and the classifier will be
[math]\displaystyle{ \ h(x) = \underset{k}{\operatorname{arg\,max}} \, (\delta_k) }[/math]
In other words we would assign each [math]\displaystyle{ \ x }[/math] to the label [math]\displaystyle{ \ k }[/math] that produces the largest [math]\displaystyle{ \ \delta_k (x) }[/math].
The reason for this becomes more obvious if we consider the components of [math]\displaystyle{ \ \delta_k (x) }[/math].
Notice that the middle component (without the coefficient), [math]\displaystyle{ \ (x-{\mathbf\mu_k})^T{\mathbf\Sigma_k}^{-1} (x-{\mathbf\mu_k}) }[/math], is expressing the squared Mahalanobis distance between [math]\displaystyle{ \ x }[/math] and [math]\displaystyle{ \ {\mathbf\mu_k} }[/math], the mean of class [math]\displaystyle{ \ k }[/math]. Therefore, by requiring that [math]\displaystyle{ \ \delta_k (x) }[/math] is maximized, we are requiring this distance to be minimized. This makes sense from a classification perspective since it is intuitive to label [math]\displaystyle{ \ x }[/math] with the class that it is closest to (on an average basis).
Also, to illustrate the role that [math]\displaystyle{ \ ln({\mathbf\pi_k}) }[/math] plays, consider the case where each class is comprised of the same number of points so that [math]\displaystyle{ \ ln({\mathbf\pi_i})\ =\ ln({\mathbf\pi_j})\ \forall i,j \in y={1,...,K} }[/math]. Then this term makes no difference in determining the appropriate label. But if a particular class [math]\displaystyle{ \ k }[/math] is comprised of many more points than others, then [math]\displaystyle{ \ ln({\mathbf\pi_k}) }[/math] is larger and increases the probability for this class (label) being assigned. This is intuitive as [math]\displaystyle{ \ \pi_k }[/math] represents the probability of a class being [math]\displaystyle{ \ k }[/math]. Therefore it must contribute to the probability that a point is in the class.
In practice when we estimate the parameters,
[math]\displaystyle{ \hat{\mathbf\pi_k}=\frac{n_k}{n},\hat{\mathbf\mu_k}=\frac{1}{n_k}\sum_{i:y_i=k}{x_i} }[/math]
[math]\displaystyle{ \hat{\mathbf\Sigma}_k=\frac{1}{n_k}\sum_{i:y_i=k}({\mathbf x}_i-{\mathbf\mu_k})({\mathbf x}_i-{\mathbf\mu_k})^T }[/math]
If we assume [math]\displaystyle{ {\mathbf\Sigma}_k={\mathbf\Sigma} }[/math] for all [math]\displaystyle{ \ k }[/math], then
[math]\displaystyle{ \hat{\mathbf\Sigma}=\frac{\sum_{r=1}^{k}n_r{\mathbf\Sigma_r}}{n} }[/math]
which is the weighted average of all [math]\displaystyle{ {\mathbf\Sigma_k} }[/math].
Fischer Discriminant Analysis (optional)
Recall that PCA finds the direction of maximum variance. Imagine PCA, MDS or Isomap as a pre-step to classification as they all reduce the dimension of the data. But if the classification is not based on the maximum variance, PCA will not work well in this case. Suppose we want to reduce a set of two-dimensional and two-class data to one dimension in order to classify them. One way to achieve this is to draw the data set in each class as close as possible (almost collapse into one point) and set these classes far apart to distinguish them. This is the intuitive interpretation of FDA. FDA does not carry the assumption of QDA and LDA, where the conditional probability distributions are a normally distributed, this allows a more flexible approach to classifying a data set.
We want to find vector [math]\displaystyle{ \!\omega }[/math] to project every point [math]\displaystyle{ \!x }[/math] to [math]\displaystyle{ \!{{\omega }^{T}}x }[/math] so that maximum variation occurs between classes. Say there are two classes 0 and 1. The points that label 0 has a mean of [math]\displaystyle{ \!{{\mu }_{0}} }[/math] and the points that label 1 has a mean of [math]\displaystyle{ \!{{\mu }_{1}} }[/math]. Then [math]\displaystyle{ {{\mu }_{0}}\to {{\omega }^{T}}{{\mu }_{0}} }[/math], and [math]\displaystyle{ {{\mu }_{1}}\to {{\omega }^{T}}{{\mu }_{1}} }[/math]
To make the two classes as far away as possible, we want to [math]\displaystyle{ \underset{\omega }{\mathop{\max }}\,||{{\omega }^{T}}{{\mu }_{0}}-{{\omega }^{T}}{{\mu }_{1}}||^2 }[/math]
In other words, [math]\displaystyle{ \underset{\omega }{\mathop{\max }}\,{{\omega }^{T}}({{\mu }_{0}}-{{\mu }_{1}}){{({{\mu }_{0}}-{{\mu }_{1}})}^{T}}\omega }[/math]
We define [math]\displaystyle{ {{S}_{B}}:=({{\mu }_{0}}-{{\mu }_{1}}){{({{\mu }_{0}}-{{\mu }_{1}})}^{T}} }[/math], which is the between-class covariance.
On the other hand, we want the variance within the same class as small as possible, we want to [math]\displaystyle{ \underset{w}{\mathop{\min }}\,{{\omega }^{T}}{{\sum }_{0}}\omega +{{\omega }^{T}}{{\sum }_{1}}\omega }[/math]
In other words, [math]\displaystyle{ \underset{w}{\mathop{\min }}\,{{\omega }^{T}}({{\sum }_{0}}+{{\sum }_{1}})\omega }[/math]
We define [math]\displaystyle{ {{S}_{W}}={{\sum }_{0}}+{{\sum }_{1}} }[/math], the within-class covariance.
Our question becomes, [math]\displaystyle{ \underset{w}{\mathop{\max }}\,=\frac{{{\omega }^{T}}{{S}_{B}}\omega }{{{\omega }^{T}}{{S}_{W}}\omega } }[/math]
Or equivalently, [math]\displaystyle{ \underset{{}}{\mathop{\max }}\,{{\omega }^{T}}{{S}_{B}}\omega }[/math] such that [math]\displaystyle{ \!{{\omega }^{T}}{{S}_{W}}\omega =1 }[/math]
Using the langrage multiplier, [math]\displaystyle{ \!L(\omega ,\lambda )={{\omega }^{T}}{{S}_{B}}\omega -\lambda ({{\omega }^{T}}{{S}_{W}}\omega -1) }[/math], we have
[math]\displaystyle{ \frac{\partial L}{\partial \omega }=0 }[/math]
[math]\displaystyle{ \Rightarrow {{S}_{B}}\omega =\lambda {{S}_{W}}\omega }[/math]
[math]\displaystyle{ \Rightarrow S_{W}^{-1}{{S}_{B}}\omega =\lambda \omega }[/math]
Therefore, [math]\displaystyle{ \!\omega }[/math] is the eigenvector of [math]\displaystyle{ S_{W}^{-1}{{S}_{B}} }[/math] (the eigenvector of the largest [math]\displaystyle{ \lambda }[/math])
Generally, FDA is a better choice than PCA in classification.