semi-supervised Learning with Deep Generative Models: Difference between revisions
Dylandrover (talk | contribs) No edit summary |
m (Conversion script moved page Semi-supervised Learning with Deep Generative Models to semi-supervised Learning with Deep Generative Models: Converting page titles to lowercase) |
||
| (28 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
= Introduction = | = Introduction = | ||
Large labelled data sets have led to massive improvements in the performance of machine learning algorithms, especially supervised neural networks. However, the world in general is not labelled and there | Large labelled data sets have led to massive improvements in the performance of machine learning algorithms, especially supervised neural networks. However, the world in general is not labelled and there exists a far greater number of unlabelled data than labelled data. A common situation is to have a comparatively small quantity of labelled data paired with a larger amount of unlabelled data. This leads to the idea of a semi-supervised learning model where the unlabelled data is used to prime the model for relevant features and the labels are then learned for classification. A prominent example of this type of model is the restricted Boltzmann machine based Deep Belief Network (DBN). Where layers of RBM are trained to learn unsupervised features of the data and then a final classification layer is applied such that labels can be assigned. | ||
Unsupervised learning | Unsupervised learning techniques sometimes create what is known as a generative model which creates a joint distribution <math>P(x, y)</math> (which can be sampled from). This is contrasted by the supervised discriminative model, which create conditional distributions <math>P(y | x)</math>. The paper combines these two methods to achieve high performance on benchmark tasks and uses deep neural networks in an innovative manner to create a layered semi-supervised classification/generation model. | ||
= Current Models and Limitations = | = Current Models and Limitations = | ||
| Line 14: | Line 14: | ||
== Latent Feature Discriminative Model (M1) == | == Latent Feature Discriminative Model (M1) == | ||
<math>p(\mathbf{z}) = \mathcal{N}(\mathbf{z}|\mathbf{0,I})</math> | The first sub-model that is described is used to model latent variables ('''z''') that embed features of the unlabelled data. Classification for this model is done separately based on the learned features from the unlabelled data. The key to this model is that the non-linear transform to capture features is a deep neural network. The generative model is based on the following equations: | ||
<div style="text-align: center;"> | |||
<math>p(\mathbf{z}) = \mathcal{N}(\mathbf{z}|\mathbf{0,I})</math> | |||
<math>p(\mathbf{x|z}) = f(\mathbf{x};\mathbf{z,\theta})</math> | <math>p(\mathbf{x|z}) = f(\mathbf{x};\mathbf{z,\theta})</math> | ||
</div> | |||
f is a likelihood function based on the parameters <math>\theta</math>. The parameters are tuned by a deep neural network. The posterior distribution <math>p(\mathbf{z}|\mathbf{x})</math> is sampled to train an arbitrary classifier for class labels <math> y </math>. Tnis approach offers substantial improvement in the performance of SVMs. | |||
== Generative Semi-Supervised Model (M2) == | |||
The second model is based on a latent variable '''z''' but the class label <math>y</math> is also treated as a latent variable and used for training. If y is available then it is used as a latent variable, but if it is not, then '''z''' is also used. The following equations describe the generative processes where <math>Cat(y|\mathbf{\pi})</math> is some multinominal distribution. f is used similarly to in M1 but with an extra parameter. Classification is treated as inference by integrating over a class of an unlabelled data sample if y is not available which is done usually with the posterior <math>p_{\theta}(y|\mathbf{x})</math>. | |||
<div style="text-align: center;"> | |||
<math>p(y) = Cat(y|\mathbf{\pi})</math> | <math>p(y) = Cat(y|\mathbf{\pi})</math> | ||
| Line 26: | Line 35: | ||
<math>p_{\theta}(\mathbf{x}|y, \mathbf{z}) = f(\mathbf{x};y,\mathbf{z,\theta})</math> | <math>p_{\theta}(\mathbf{x}|y, \mathbf{z}) = f(\mathbf{x};y,\mathbf{z,\theta})</math> | ||
</div> | |||
Another way to see this model is as a hybrid continuous-discrete mixture model, where the parameters are shared between the different components of the mixture. | |||
== Stacked Generative Semi-Supervised Model (M1+M2) == | == Stacked Generative Semi-Supervised Model (M1+M2) == | ||
The two aforementioned models are concatenated to form the final model. The method in which this works is that M1 is learned while M2 uses the latent variables from model M1 ('''z_1''') as the data as opposed to raw values '''x'''. The following equations describe the entire model. The distributions of <math>p_{\theta}(\mathbf{z1}|y,\mathbf{z2})</math> and <math>p_{\theta}(\mathbf{x|z1})</math> are parametrized as deep neural networks. | |||
<div style="text-align: center;"> | |||
<math>p_{\theta}(\mathbf{x}, y, \mathbf{z1, z2}) = p(y)p(\mathbf{z2})p_{\theta}(\mathbf{z1}|y, \mathbf{z2})p_{\theta}(\mathbf{x|z1})</math> | <math>p_{\theta}(\mathbf{x}, y, \mathbf{z1, z2}) = p(y)p(\mathbf{z2})p_{\theta}(\mathbf{z1}|y, \mathbf{z2})p_{\theta}(\mathbf{x|z1})</math> | ||
[[File:Kingma_2014_1.png |thumb|upright=3]] | </div> | ||
The problems of intractable posterior distributions is solved with the work of Kingma and Welling using variational inference. These inference networks are not described in detail in the paper. The following algorithms show the method in which the optimization for the methods is performed. | |||
[[File:Kingma_2014_1.png |centre|thumb|upright=3|]] | |||
The posterior distributions are, as usual, intractable, but this problem is resolved through the use of a fixed form distribution <math>q_{\phi}(\mathbf{x|z}</math>, with <math>\phi</math> as parameters that approximate <math>p(\mathbf{z|x})</math>. The equation <math>q_{\phi}</math> is created as an inference network, which allows for the computation of global parameters and does not require computation for each individual data point. | |||
[[File:kingma_2014_4.png |centre|]] | |||
In the equations, <math>\,\sigma_{\phi}(x)</math> is a vector of standard deviations <math>\,\pi_{\theta}(x)</math> is a probability vector, and the functions <math>\,\mu_{\phi}(x), \sigma_{\phi}(x) </math> and <math> \,\pi_{\theta}(x)</math> are treated as MLPs for optimization. | |||
The above algorithm is not more computationally expensive than approaches based on autoencoders or neural models, and has the advantage of being fully probabilistic. The complexity of a single joint update of M<sub>1</sub> can be written as C<sub>M1</sub> = MSC<sub>MLP</sub> where M is the batch size, S is the number of samples of ε and C<sub>MLP</sub> has the form O(KD<sup>2</sup>), where K is the number of layers in the model and D is the average dimension of the layers. The complexity for M<sub>2</sub> has the form LC<sub>M1</sub> where L is the number of labels. All above models can be trained with any of EM algorithm, stochastic gradient variational Bayes, or stochastic backpropagation methods. | |||
= Results = | = Results = | ||
The complexity of M1 can be estimated by using the complexity of the MLP used for the parameters which is equal to <math>C_{MLP} = O(KD^2)</math> with K is the number of layers and D is the average of the neurons in each layer of the network. The total complexity is <math>C_{M1}=MSC_{MLP}</math> with M = size of the mini-batch and S is the number of samples. Similarly the complexity of M2 is <math>C_{M2}=LC_{M1}</math>, where L is the number of labels. Therefore the combined complexity of the model is just a combination of these two complexities. This is equivalent to the lowest complexities of similar models, however, this approach achieves better results as seen in the following table. | The complexity of M1 can be estimated by using the complexity of the MLP used for the parameters which is equal to <math>C_{MLP} = O(KD^2)</math> with K is the number of layers and D is the average of the neurons in each layer of the network. The total complexity is <math>C_{M1}=MSC_{MLP}</math> with M = size of the mini-batch and S is the number of samples. Similarly the complexity of M2 is <math>C_{M2}=LC_{M1}</math>, where L is the number of labels. Therefore the combined complexity of the model is just a combination of these two complexities. This is equivalent to the lowest complexities of similar models, however, this approach achieves better results as seen in the following table. | ||
The results are better across all labelled set sizes for the M1+M2 model and drastically better for when the number of labelled data samples is very small (100 out of 50000). | |||
[[File:kingma_2014_2.png | centre|]] | [[File:kingma_2014_2.png | centre|]] | ||
The following figure demonstrates the model's ability to generate images through conditional generation. The class label was fixed and then the latent variables, '''z''', were altered. The figure shows how the latent variables were varied and how the generated digits are similar for similar values of '''z'''s. Parts b and c of the figure use a test image to generate images that belong to a similar set of '''z''' values (images that are similar). | |||
[[File:kingma_2014_3.png |thumb|upright=3|centre|]] | |||
[ | A commendable part of this paper is that they have actually included their [http://github.com/dpkingma/nips14-ssl source code]. | ||
= Conclusions and Critique = | = Conclusions and Critique = | ||
The results using this method are obviously impressive and the fact that the model achieves this with comparable computation times compared to the other models is notable. The heavy use of approximate inference methods shows great promise in improving generative models and thus semi-supervised methods. The authors discuss the potential of combining this method with the supervised methods that have given state-of-the-art results in image processing, convolutional neural networks. This might be possible as all parameters in their models are optimized using neural networks. The final model acts as a approximate Bayesian inference model. | |||
The architecture of the model is not very explicit in the paper, that is, a diagram showing the layout of the entire model would have ameliorated understanding. Another weak point is that they fail to compare their method to existing tractable inference neural network methods. There is no comparison to Sum Product Networks nor Deep Belief Networks. | |||
Latest revision as of 09:46, 30 August 2017
Introduction
Large labelled data sets have led to massive improvements in the performance of machine learning algorithms, especially supervised neural networks. However, the world in general is not labelled and there exists a far greater number of unlabelled data than labelled data. A common situation is to have a comparatively small quantity of labelled data paired with a larger amount of unlabelled data. This leads to the idea of a semi-supervised learning model where the unlabelled data is used to prime the model for relevant features and the labels are then learned for classification. A prominent example of this type of model is the restricted Boltzmann machine based Deep Belief Network (DBN). Where layers of RBM are trained to learn unsupervised features of the data and then a final classification layer is applied such that labels can be assigned. Unsupervised learning techniques sometimes create what is known as a generative model which creates a joint distribution [math]\displaystyle{ P(x, y) }[/math] (which can be sampled from). This is contrasted by the supervised discriminative model, which create conditional distributions [math]\displaystyle{ P(y | x) }[/math]. The paper combines these two methods to achieve high performance on benchmark tasks and uses deep neural networks in an innovative manner to create a layered semi-supervised classification/generation model.
Current Models and Limitations
The paper claims that existing unlabelled data models do not scale well for very large sets of unlabelled data. One example that they discuss is the Transductive SVM, which they claim does not scale well and that optimization for them is a problem. Graph based models suffer from sensitivity to their graphical structure which may make them rigid. Finally they briefly discuss other neural network based methods such as the Manifold Tangent Classifier that uses contrastive auto-encoders (CAEs) to deduce the manifold on which data lies. Based on the manifold hypothesis this means that similar data should not lie far from the manifold and they then can use something called TangentProp to train a classifier based on the manifold of the data.
Proposed Method
Rather than use the methods mentioned above the team suggests that using generative models based on neural networks would be beneficial. Current generative models lack string inference and scalability though. The paper proposes a method that uses variational inference for semi-supervised classification that will employ deep neural networks.
Latent Feature Discriminative Model (M1)
The first sub-model that is described is used to model latent variables (z) that embed features of the unlabelled data. Classification for this model is done separately based on the learned features from the unlabelled data. The key to this model is that the non-linear transform to capture features is a deep neural network. The generative model is based on the following equations:
[math]\displaystyle{ p(\mathbf{z}) = \mathcal{N}(\mathbf{z}|\mathbf{0,I}) }[/math]
[math]\displaystyle{ p(\mathbf{x|z}) = f(\mathbf{x};\mathbf{z,\theta}) }[/math]
f is a likelihood function based on the parameters [math]\displaystyle{ \theta }[/math]. The parameters are tuned by a deep neural network. The posterior distribution [math]\displaystyle{ p(\mathbf{z}|\mathbf{x}) }[/math] is sampled to train an arbitrary classifier for class labels [math]\displaystyle{ y }[/math]. Tnis approach offers substantial improvement in the performance of SVMs.
Generative Semi-Supervised Model (M2)
The second model is based on a latent variable z but the class label [math]\displaystyle{ y }[/math] is also treated as a latent variable and used for training. If y is available then it is used as a latent variable, but if it is not, then z is also used. The following equations describe the generative processes where [math]\displaystyle{ Cat(y|\mathbf{\pi}) }[/math] is some multinominal distribution. f is used similarly to in M1 but with an extra parameter. Classification is treated as inference by integrating over a class of an unlabelled data sample if y is not available which is done usually with the posterior [math]\displaystyle{ p_{\theta}(y|\mathbf{x}) }[/math].
[math]\displaystyle{ p(y) = Cat(y|\mathbf{\pi}) }[/math]
[math]\displaystyle{ p(\mathbf{z}) = \mathcal{N}(\mathbf{z}|\mathbf{0,I}) }[/math]
[math]\displaystyle{ p_{\theta}(\mathbf{x}|y, \mathbf{z}) = f(\mathbf{x};y,\mathbf{z,\theta}) }[/math]
Another way to see this model is as a hybrid continuous-discrete mixture model, where the parameters are shared between the different components of the mixture.
Stacked Generative Semi-Supervised Model (M1+M2)
The two aforementioned models are concatenated to form the final model. The method in which this works is that M1 is learned while M2 uses the latent variables from model M1 (z_1) as the data as opposed to raw values x. The following equations describe the entire model. The distributions of [math]\displaystyle{ p_{\theta}(\mathbf{z1}|y,\mathbf{z2}) }[/math] and [math]\displaystyle{ p_{\theta}(\mathbf{x|z1}) }[/math] are parametrized as deep neural networks.
[math]\displaystyle{ p_{\theta}(\mathbf{x}, y, \mathbf{z1, z2}) = p(y)p(\mathbf{z2})p_{\theta}(\mathbf{z1}|y, \mathbf{z2})p_{\theta}(\mathbf{x|z1}) }[/math]
The problems of intractable posterior distributions is solved with the work of Kingma and Welling using variational inference. These inference networks are not described in detail in the paper. The following algorithms show the method in which the optimization for the methods is performed.
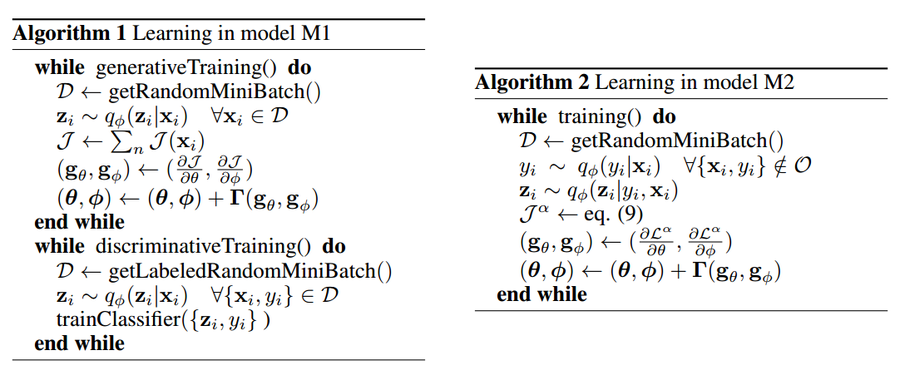
The posterior distributions are, as usual, intractable, but this problem is resolved through the use of a fixed form distribution [math]\displaystyle{ q_{\phi}(\mathbf{x|z} }[/math], with [math]\displaystyle{ \phi }[/math] as parameters that approximate [math]\displaystyle{ p(\mathbf{z|x}) }[/math]. The equation [math]\displaystyle{ q_{\phi} }[/math] is created as an inference network, which allows for the computation of global parameters and does not require computation for each individual data point.
In the equations, [math]\displaystyle{ \,\sigma_{\phi}(x) }[/math] is a vector of standard deviations [math]\displaystyle{ \,\pi_{\theta}(x) }[/math] is a probability vector, and the functions [math]\displaystyle{ \,\mu_{\phi}(x), \sigma_{\phi}(x) }[/math] and [math]\displaystyle{ \,\pi_{\theta}(x) }[/math] are treated as MLPs for optimization.
The above algorithm is not more computationally expensive than approaches based on autoencoders or neural models, and has the advantage of being fully probabilistic. The complexity of a single joint update of M1 can be written as CM1 = MSCMLP where M is the batch size, S is the number of samples of ε and CMLP has the form O(KD2), where K is the number of layers in the model and D is the average dimension of the layers. The complexity for M2 has the form LCM1 where L is the number of labels. All above models can be trained with any of EM algorithm, stochastic gradient variational Bayes, or stochastic backpropagation methods.
Results
The complexity of M1 can be estimated by using the complexity of the MLP used for the parameters which is equal to [math]\displaystyle{ C_{MLP} = O(KD^2) }[/math] with K is the number of layers and D is the average of the neurons in each layer of the network. The total complexity is [math]\displaystyle{ C_{M1}=MSC_{MLP} }[/math] with M = size of the mini-batch and S is the number of samples. Similarly the complexity of M2 is [math]\displaystyle{ C_{M2}=LC_{M1} }[/math], where L is the number of labels. Therefore the combined complexity of the model is just a combination of these two complexities. This is equivalent to the lowest complexities of similar models, however, this approach achieves better results as seen in the following table.
The results are better across all labelled set sizes for the M1+M2 model and drastically better for when the number of labelled data samples is very small (100 out of 50000).
The following figure demonstrates the model's ability to generate images through conditional generation. The class label was fixed and then the latent variables, z, were altered. The figure shows how the latent variables were varied and how the generated digits are similar for similar values of zs. Parts b and c of the figure use a test image to generate images that belong to a similar set of z values (images that are similar).
A commendable part of this paper is that they have actually included their source code.
Conclusions and Critique
The results using this method are obviously impressive and the fact that the model achieves this with comparable computation times compared to the other models is notable. The heavy use of approximate inference methods shows great promise in improving generative models and thus semi-supervised methods. The authors discuss the potential of combining this method with the supervised methods that have given state-of-the-art results in image processing, convolutional neural networks. This might be possible as all parameters in their models are optimized using neural networks. The final model acts as a approximate Bayesian inference model.
The architecture of the model is not very explicit in the paper, that is, a diagram showing the layout of the entire model would have ameliorated understanding. Another weak point is that they fail to compare their method to existing tractable inference neural network methods. There is no comparison to Sum Product Networks nor Deep Belief Networks.