SPINS Tutorial: Difference between revisions
| (38 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
This is a tutorial for getting up to speed with SPINS using the case <code>mode1_mode2</code>. The first part of the tutorial discusses running the case with the default parameters, some basic analysis tools and techniques, and manually changing parameters. The second part of the tutorial explores more parameter combinations using Matlab driver scripts to automate the workflow. The final part | This is a tutorial for getting up to speed with SPINS using the case <code>mode1_mode2</code>. The first part of the tutorial discusses running the case with the default parameters, some basic analysis tools and techniques, and manually changing parameters. The second part of the tutorial explores more parameter combinations using Matlab driver scripts to automate the workflow. The final part uses a Matlab script <code>spins_refinement.m</code> to interpolate an existing SPINS output onto a finer grid and restart SPINS at higher resolution. | ||
You must first have followed the [[SPINS User Guide#How to get SPINS running|instructions]] to install SPINS and compile the case file. | You must first have followed the [[SPINS User Guide#How to get SPINS running|instructions]] to install SPINS and compile the case file. | ||
| Line 17: | Line 17: | ||
# Diagnostic files | # Diagnostic files | ||
#* These will generally be text files and contain information about the simulation | #* These will generally be text files and contain information about the simulation | ||
#* Matlab plotters for the diagnostic files are found in | #* Matlab plotters for the diagnostic files are found in [[SPINS MATLAB tools]], available [https://github.com/ddeepwel/SPINSmatlab here]. | ||
{| class="wikitable" | {| class="wikitable" | ||
!colspan="4"|Diagnostic Files | !colspan="4"|Diagnostic Files | ||
| Line 45: | Line 45: | ||
Matlab and Python are the primary languages used for analyzing SPINS simulations. These are great for computation of quantities specific to your study. The [https://git.uwaterloo.ca/bastorer/SPINSpy Python (SPINSpy)] and [https://git.uwaterloo.ca/ddeepwel/SPINSmatlab Matlab (SPINSmatlab)] packages contain much of the important tools for each language. Three dimensional visualization should use Paraview or VisIt (see [[Visualization]]). | Matlab and Python are the primary languages used for analyzing SPINS simulations. These are great for computation of quantities specific to your study. The [https://git.uwaterloo.ca/bastorer/SPINSpy Python (SPINSpy)] and [https://git.uwaterloo.ca/ddeepwel/SPINSmatlab Matlab (SPINSmatlab)] packages contain much of the important tools for each language. Three dimensional visualization should use Paraview or VisIt (see [[Visualization]]). | ||
Here, we will use Matlab to make a simple plot of the density field. We assume that the SPINSmatlab package is on your path | Here, we will use Matlab to make a simple plot of the density field. We assume that the SPINSmatlab package is on your path. | ||
#<code>gdpar = spins_gridparams(); split_gdpar</code> | #<code>gdpar = spins_gridparams(); split_gdpar</code> | ||
| Line 119: | Line 119: | ||
Try it yourself. See what each parameter does when they're changed. | Try it yourself. See what each parameter does when they're changed. | ||
= | = Extend to 3D = | ||
= Part 2: Matlab driver scripts = | Often, a case initially simulated in 2D will undergo various physical processes that allow it to become 3D, thus, in many circumstances, a 2D simulation could be a misrepresentation of the overall dynamics. To this end, there are different mechanisms built into SPINS to deal with this. The simplest process is the run a 2D simulation until you believe that 3D effects will have occurred. This is problem specific and is left up to the choice of the modeller. | ||
Once a particular output is chosen, the simulation can be extended into the spanwise (assumed to be the <math> y </math> dimension), and the simulation can be restarted from that point and run in 3D. | |||
To use this functionality, the script <code> extend_all()</code> has been included in the <code>SPINSmatlab</code> toolbox. | |||
# Run the simulation until the output 30 | |||
# Navigate to the directory where the output is stored. Go into MATLAB and enter <code>extend_all(30,0.1,32)</code>. | |||
# <code>extend_all</code> creates a sub-directory called <code>extended/</code> which contains the now 3D data, the <code>spins.conf</code> with updated grid and restarting information, and the executable. | |||
# Navigate into the <code>extended/</code> directory and run the executable. | |||
Note now that the data is three dimensional, to read it into MATLAB, you must choose a slice (an <math>x-y</math>, <math>x-z</math>, or <math>y-z</math> cross-section) to plot. This is commonly done in two ways. | |||
# You can use <code>spins_reader()</code> to read in slices of fields and grids by entering <code>spins_reader(field,output,x_start:x_end,y_start:y_end,z_start:z_end)</code>. <code>[]</code> implies the entire dimension. This is imperative for large datasets, as MATLAB has immense difficulty opening very large arrays without freezing. | |||
# For smaller sized 3D data sets, the entire field can be loaded in and slices taken at different points in analysis. To get a fast plot, use the following command: | |||
imagesc(flipud(squeeze(u(:,y_ind,:))')) | |||
if you want to plot an <math>x-z</math> slice at the span corresponding to <code>y_ind</code>. The <code>squeeze()</code> command remove the singleton dimension leaving the field as a 2D array. The combination of the <code>flipud()</code> and transpose (<code>'</code>) operations allow the bottom of the field to align with the bottom of the plot. | |||
Note that an <math>x-y</math> slice does not need the <code>flipud()</code> and transpose (<code>'</code>) operations. | |||
= Editing Casefiles = | |||
This needs to be done. | |||
<!-- NOTE THAT THE SECTIONS BELOW NEED TO BE REFINED IF THEY ARE TO BE INLCUDED. CONSIDER ADDING THEM AS A SEPARATE WIKI PAGE AS THEY ARE NOT 'NEED-TO-KNOW' ASPECTS OF SPINS. THIS PAGE IS RESERVED FOR THE MOST BASIC SPINS FUNCTIONALITY --> | |||
<!-- | |||
= Part 2: Matlab driver scripts (UNDER CONSTRUCTION)= | |||
Part 2 and 3 extend the previous tutorial. In the remaining sections of this tutorial you will: | Part 2 and 3 extend the previous tutorial. In the remaining sections of this tutorial you will: | ||
| Line 129: | Line 156: | ||
== Setup == | == Setup == | ||
To get started on these parts, clone the [https://git.uwaterloo.ca/tghill/SPINS-refinement/ SPINS refinement git repo]. | To get started on these parts, clone the [https://git.uwaterloo.ca/tghill/SPINS-refinement/ SPINS refinement git repo]. | ||
This will clone a directory called <code>SPINS-refinement/</code>. Included in this directory are several subdirectories. Relevant to this part of the tutorial is the directory called <code>tutorial/</code>. Navigate to this directory. | |||
<code>tutorial/</code> contains all the files needed to run an already designed selection of cases with no modification necessary. | |||
Included within the directory are several scripts of interest. | |||
First look at <code>mode1_mode2.txt</code>. In this file are the different casenames for the parameter sweep, including the parameters which are varied. <code>casenames</code> in this text file must match <code>casenames</code> in the <code>matlab.sh</code> submission script! | |||
The matlab script <code>driver_mode1_mode2.m</code> sets up the cases. It creates directories for each case (named according to the casename), writes the parameters to a new <code>spins.conf</code> file, and creates the initial u, v, w, and rho files. This script has its own documentation on the git repository, and the code is well commented. | |||
== Run == | == Run == | ||
| Line 144: | Line 177: | ||
# lamp. This cases increases the height and width of the mode 1 and mode 2 regions to create larger amplitude waves. The pycnocline is also set to be in the middle of the tank. | # lamp. This cases increases the height and width of the mode 1 and mode 2 regions to create larger amplitude waves. The pycnocline is also set to be in the middle of the tank. | ||
Now we actually run these cases. | Now we actually run these cases. | ||
To do this, submit the <code>matlab.sh</code> script to the scheduler (<code>sbatch matlab.sh</code>). This job runs <code>wavereader.x</code> and submits a job for each case. Specifically, it submits the <code>submit.sh</code> script from each case directory. | |||
== Designing your own cases == | |||
You can add your own cases by adding them to the text file <code>mode1_mode2.txt</code>. The casenames in this text file must match the casenames in the <code>matlab.sh</code> submission script! Try adding a case to the existing list and running all the cases again. | |||
Modifying parameters that aren't already a column in the text file is somewhat more work. This can be done in 3 steps below. | |||
# Add the parameter as a column to the text file listing the cases. As a concrete example, we will add a column changing the viscosity. <code>casename, Lx, Lz, Nx, Nz, H1, H2, L1, L2, pyc_loc, visco</code> | |||
# In the file <code>driver_mode1_mode2.m</code> remove the line <code>visco=1.e-6</code>. This is not strictly necessary, but should be done anyways to avoid possible confusion later. | |||
# Within this file, add the line <code>visco=par.visco(numcase);</code> under the header <code>%stratification</code>. The idea here is that you need to tell the driver script to read this new column in the table and assign the relevant parameter a new value for each case. | |||
Further documentation and examples are available on the SPINS-refinement repository. | |||
== | == Summary == | ||
For future reference, the user may want to refer to the following flow chart as a reminder of how to use the workflow functionality. | |||
[[File:workflow.png|frameless|border|caption]] | |||
= Part 3: Refining the grid from existing outputs = | = Part 3: Refining the grid from existing outputs (UNDER CONSTRUCTION)= | ||
Say you ran a simulation and noticed that after a certain time, the model output began to exhibit signs of small unphysical oscillations. Or perhaps you almost managed to resolve a feature of interest near the end of your simulation. Both of these scenarios could be solved by restarting your simulation from the beginning with higher resolution, however, in the interest of time (especially for large runs), it may be useful to start from an earlier point in your existing simulation with a higher resolution. | Say you ran a simulation and noticed that after a certain time, the model output began to exhibit signs of small unphysical oscillations. Or perhaps you almost managed to resolve a feature of interest near the end of your simulation. Both of these scenarios could be solved by restarting your simulation from the beginning with higher resolution, however, in the interest of time (especially for large runs), it may be useful to start from an earlier point in your existing simulation with a higher resolution. | ||
This is where the script <code>spins_refinement.m</code> becomes useful. <code>spins_refinement.m</code> takes all the | This is where the script <code>spins_refinement.m</code> becomes useful. <code>spins_refinement.m</code> takes all the fields at a specified output, makes a finer grid, and interpolates the existing outputs onto this finer grid. It then outputs new fields that can be read by <code>wave_reader.x</code>. This effectively allows you to restart a simulation from an earlier output but with higher resolution. | ||
Spectral refinement is also available, more information below. | |||
== Usage == | == Usage == | ||
Using <code>spins_refinement.m</code> is easy (trust us). There are a few steps that must be taken before using it though. | |||
First, <code>wave_reader.x</code> must be compiled on the machine you intend to run the refined cases on. The process for this is identical to the process for compiling <code>mode1_mode2.x</code> above, except you simply compile <code>wave_reader.cpp</code>. Once this is complete, the next step is to clone the <code>SPINSMatlab</code> functions. The process for this is introduced earlier in this tutorial. | |||
The necessary steps to use <code>spins_refinement.m</code> as well as the code snippet are given below. | |||
nx = 4; % nx times of original resolution in x | |||
nz = 2; % nz times of original resolution in z | |||
ii = 100; % number of output to interpolate | |||
method = 'nearest'; % Matlab built-in interpolation method | |||
% 'nearest', 'linear', 'spline' or 'cubic' | |||
# The first step is to adjust the parameters in the script that specify the new grid, the first refined output, and the interpolation method. The only parameters that the user should have to edit are given above.<br /> The parameters <code>nx = 4</code> and <code>nz = 2</code> mean that the number of grid points in the horizontal and vertical directions will be increased by a factor of 4 and 2 respectively. | |||
# Next, the line <code>ii = 100</code> determined what output the user would like to refine and begin the new simulation from. This output will be the new initial condition for the higher resolution version of your case. Be aware that the output you choose must be sufficiently early that any transient motion due to the interpolation can be removed by viscosity or filtering before the motion of interest is analyzed. | |||
# Lastly, you may also edit the variable ‘method’, which changes the interpolation method used. Currently, the only methods available are `nearest', `linear', `spline', or `cubic'. In the future, a separate script for spectral interpolation may be added. | |||
# With the new grid parameters specified, move <code>spins_reader.m</code> and <code>spins_refinement.m</code> into the working directory and run <code>spins_refinement.m</code>. The script will create the interpolated fields on the finer grid and place them in a new subdirectory called <code>high_res</code> along with a new <code>spins.conf</code>. | |||
# Lastly, copy the submit script and <code>wave_reader.x</code> into the directory <code>high_res</code> and submit the refined case. | |||
Spectral refinement (rather than the matlab interpolation schemes) is available in [[SPINS resize]] (a subset of the [https://git.uwaterloo.ca/ddeepwel/SPINSmatlab SPINSmatlab toolbox]). This refinement toolbox also allows for arbitrary refinement or coarsening, rather than a mutliplication of the current resolution. | |||
== Summary == | |||
For future reference, the user may want to refer to the following flow chart as a reminder of how to use the <code>spins_refinement.m</code> functionality. | |||
[[File:refinement.png|frameless|border|caption]] | |||
--> | |||
Latest revision as of 11:13, 18 October 2021
This is a tutorial for getting up to speed with SPINS using the case mode1_mode2. The first part of the tutorial discusses running the case with the default parameters, some basic analysis tools and techniques, and manually changing parameters. The second part of the tutorial explores more parameter combinations using Matlab driver scripts to automate the workflow. The final part uses a Matlab script spins_refinement.m to interpolate an existing SPINS output onto a finer grid and restart SPINS at higher resolution.
You must first have followed the instructions to install SPINS and compile the case file.
Part 1: Getting familiar with SPINS
Run
Transfer the executable (mode1_mode2.x) and the configuration file (spins.conf) into a clean directory.
Run simulation by executing mpirun -np 4 ./mode1_mode2.x on the command line, or submit the job to the queue on a Compute Canada system with a submit script (see submitting jobs to Graham).
This will create two types of files
- Output files
- These are of the form
rho.3where the name (rho) is the variable and the extension is the output number
- - For example rho.3 is the density field at output number 3. Zero-indexing is used, so this is the 4th density file
- Since these can be memory expensive, they are created relatively infrequently
- These are of the form
- Diagnostic files
- These will generally be text files and contain information about the simulation
- Matlab plotters for the diagnostic files are found in SPINS MATLAB tools, available here.
| Diagnostic Files | |||
|---|---|---|---|
| File | Optional | Matlab plotter | Description |
| diagnostics.txt | No | plot_diagnos | High temporal resolution information about the simulation (occurs at every time step). Examples include: KE, max velocity, energy budget terms, simulation timings. |
| stresses_top.txt and/or stresses_bottom.txt | Yes | plot_stress | Surface forces and surface stress extrema at the top/bottom surfaces. Only applicable if no slip boundary condition is used. |
| plot_times.txt | No | plot_diagnos | Explicit statement of the time associated with each output. Also gives the time to write output files. |
Analyze
Matlab and Python are the primary languages used for analyzing SPINS simulations. These are great for computation of quantities specific to your study. The Python (SPINSpy) and Matlab (SPINSmatlab) packages contain much of the important tools for each language. Three dimensional visualization should use Paraview or VisIt (see Visualization).
Here, we will use Matlab to make a simple plot of the density field. We assume that the SPINSmatlab package is on your path.
gdpar = spins_gridparams(); split_gdpar- This reads in the grid and parameters and places them in a structure (
gdpar).split_gdparseparates the two sub-structures into the structuresparandgd, which are the parameters and grid, respectively. parcontains information fromspins.confin addition to further simulation parametersgdcontains the grid. x,y,z grids can be extracted withgd.x,gd.y,orgd.z- The default setting is that the grid is unmapped and vector grids are produced, to get the full grid use
gdpar = spins_gridparams('FastFull');orgdpar = spins_gridparams('Full');
- This reads in the grid and parameters and places them in a structure (
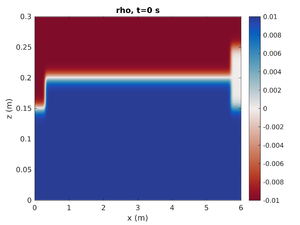
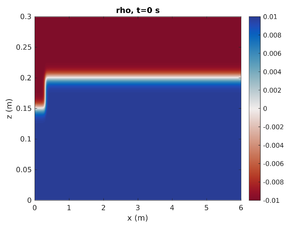
spins_plot2d('rho',0);- First argument is the field to plot
- Second argument is the output number. The simulation time can also be specified in this argument as a string.
spins_plot2d('rho','2');will plot the simulation at the closest output to t=2s. Many optional arguments exist (typehelp spins_plot2dfor more options).
To read an individual file:
u = spins_reader('u',10);
Restart
Should you need to restart the simulation (due to expenditure of allocated time, require more time, a node failure, or otherwise) simply change the restart flags in the configuration file.
The simplest change is to set
restart = true restart_time = 5.5 restart_sequence = 11
where the simulation will restart at output 11 which corresponds to t=5.5s. These numbers can be found in the last row of plot_times.txt.
SPINS also has an automatic safety dump if the allocated time is close to expiring (See automatically specify the runtime for more info). In this case, if the safety write was done successfully then you only need to set
restart_from_dump = true
Change
If after analyzing the simulation you realize that you wanted to change a parameter (it wasn't quite right), then you only need to change the parameter in the configuration file (as opposed to older editions of SPINS where you'd need to recompile the entire code).
Here is a description of the problem parameters:
--delta_rho arg Density difference between upper and lower layers (as a percentage of rho_0) --pyc_loc arg Pycnocline location --h_halfwidth arg Pycnocline half-width --delta_x arg Horizontal transition half-width --H2 arg Height of mixed region (mode-2) - sets mode-2 wave amplitude --H1 arg Heigth of mixed region (mode-1) - sets mode-1 wave amplitude --L1 arg Length of region 1 (mode-2) --L2 arg Length of region 2 (mode-1) --dye1_loc arg Location of dye 1 --dye2_loc arg Location of dye 2 --dye1_width arg Width of dye 1 --dye2_width arg Width of dye 2 --dye_halfwidth arg Sharpness of the dye transition --enable_tracer Enable evolution of second tracers
If you run the simulation in the same directory, remember to clear all simulation files (output and diagnostic files).
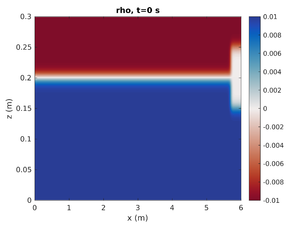
Only Mode-1 ISW
To change the simulation to only have the left mode-1 ISW change spins.conf to have:
H2 = 0.0
This will set the mode-2 amplitude to zero.
Only Mode-2 ISW
To change the simulation to only have the right mode-2 ISW change spins.conf to have:
H1 = 0.0
This will set the mode-1 amplitude to zero.
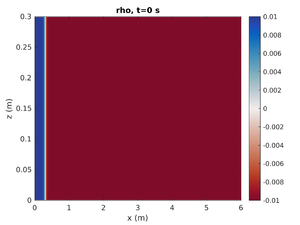
Gravity Current
To change the simulation to have a rightward moving gravity current change spins.conf to have:
pyc_loc = -0.2 H1 = -1.0 H2 = 0.0
This will move the pycnocline below the bottom boundary, set the mode-1 amplitude to result in a region of high density at on end of the tank, and give no mode-2 contribution.
Try it yourself. See what each parameter does when they're changed.
Extend to 3D
Often, a case initially simulated in 2D will undergo various physical processes that allow it to become 3D, thus, in many circumstances, a 2D simulation could be a misrepresentation of the overall dynamics. To this end, there are different mechanisms built into SPINS to deal with this. The simplest process is the run a 2D simulation until you believe that 3D effects will have occurred. This is problem specific and is left up to the choice of the modeller.
Once a particular output is chosen, the simulation can be extended into the spanwise (assumed to be the dimension), and the simulation can be restarted from that point and run in 3D.
To use this functionality, the script extend_all() has been included in the SPINSmatlab toolbox.
- Run the simulation until the output 30
- Navigate to the directory where the output is stored. Go into MATLAB and enter
extend_all(30,0.1,32). extend_allcreates a sub-directory calledextended/which contains the now 3D data, thespins.confwith updated grid and restarting information, and the executable.- Navigate into the
extended/directory and run the executable.
Note now that the data is three dimensional, to read it into MATLAB, you must choose a slice (an , , or cross-section) to plot. This is commonly done in two ways.
- You can use
spins_reader()to read in slices of fields and grids by enteringspins_reader(field,output,x_start:x_end,y_start:y_end,z_start:z_end).[]implies the entire dimension. This is imperative for large datasets, as MATLAB has immense difficulty opening very large arrays without freezing.
- For smaller sized 3D data sets, the entire field can be loaded in and slices taken at different points in analysis. To get a fast plot, use the following command:
imagesc(flipud(squeeze(u(:,y_ind,:))'))
if you want to plot an slice at the span corresponding to y_ind. The squeeze() command remove the singleton dimension leaving the field as a 2D array. The combination of the flipud() and transpose (') operations allow the bottom of the field to align with the bottom of the plot.
Note that an slice does not need the flipud() and transpose (') operations.
Editing Casefiles
This needs to be done.